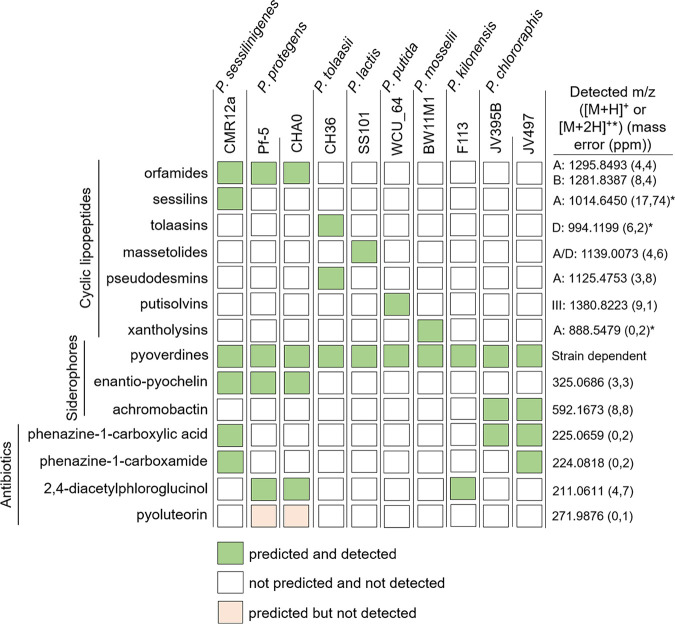
FIG 1.
Predicted and detected bioactive secondary metabolites (BSMs) produced by the Pseudomonas strains used in this study. Metabolites with biosynthetic gene clusters (BGCs) predicted by antiSMASH 6.0 (42) and detected ([M+H]+ or [M+2H]+, the latter is indicated with an asterisk) by UPLC-qTOF MS in crude cell-free supernatant after growth in CAA medium are represented by green squares, while undetected BSMs are represented by light red squares. Detected m/z and mass errors in parts per million (based on one measurement from one strain [orfamides, CMR12a; achromobactin, JV497; phenazines, CMR12a; 2,4-diacetylphloroglucinol, Pf-5]) corresponding to the main variant(s) (indicated with the letters) detected are presented. The data are representative of the two independent repetitions.