Figure 5. RGG domains interact with RNA polymerase II C-terminal heptad tail.
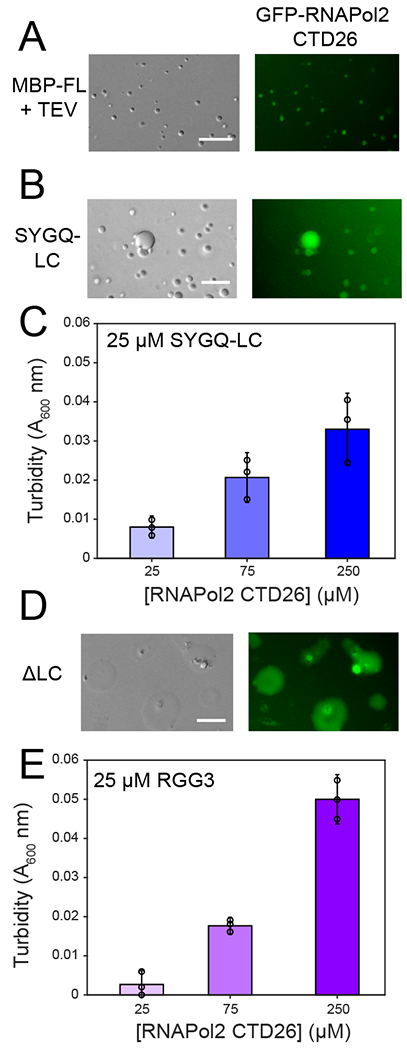
A) Recruitment of GFP-CTD26 into droplets formed by 5 μM MBP-FUS FL after cleavage of the N-terminal MBP solubility tag by addition of TEV protease. For all panels in this figure, micrographs are from one representative experiment repeated two times. B) Recruitment of GFP-RNAPol II CTD26 into droplets formed by 300 μM SYGQ-LC. C) Turbidity of 25 μM SYGQ-LC in the presence of increasing concentrations of untagged RNA polymerase II CTD. The data subtract turbidity values of control SYGQ-LC alone. Data are plotted as mean ± s.d. from n=3 replicates in one representative data set out of two independent experiments. D) Recruitment of GFP-CTD26 into droplets formed by 50 μM ΔSYGQ LC. E) Turbidity of 25 μM RGG3 in the presence of increasing concentrations of untagged RNA polymerase II CTD. The data subtract turbidity values of control RGG3 alone. Data are plotted as mean ± s.d. from n=3 replicates in one representative data set out of two independent experiments. For all micrographs in this figure, data shown are from one representative dataset of two experiments.
