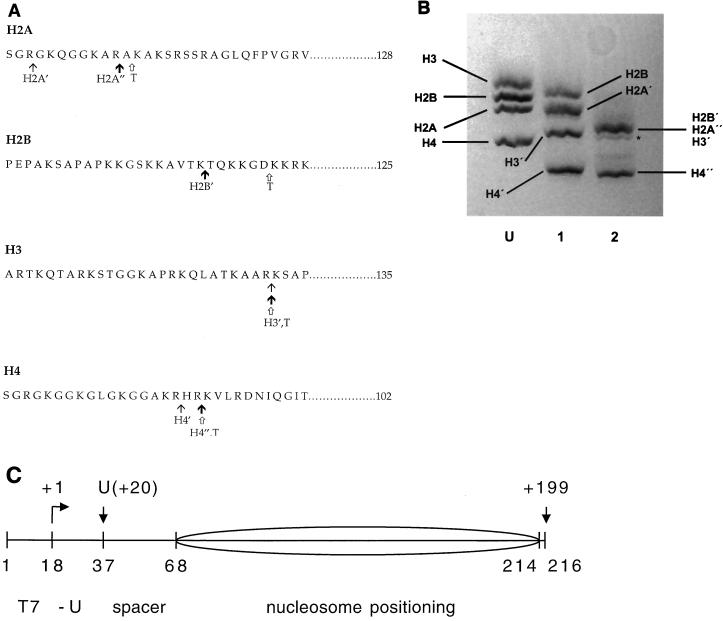
FIG. 1.
Protease cleavage of the core histone tail domains. (A) Amino acid sequences of the four chicken erythrocyte core histones showing the locations of the cleavage sites for limited or more-extensive clostripain digestion (see reference 8) and trypsin digestion (see reference 5). The locations of the clostripain cleavage sites shown are those determined in past studies on rat histones (8), which, based on the high degree of amino acid sequence conservation between the two organisms and the close similarity of SDS-PAGE analyses of the time courses of digestion for the two cases, are presumed to be identical. The tet-no-tails sample obtained by limited clostripain digestion (at points indicated by thin arrows, with prime designations on the histones) contains slightly shortened H2A′ (lacking 3 residues from the N terminus), undigested H2B, tailless H3′ (cleaved after residues 26), and tailless H4′ (cleaved after residue 17). The oct-no-tails sample obtained by more-extensive clostripain digestion (at points indicated by bold arrows, with prime or double-prime designations on the histones) contains H2A" (which differs from H2A′ by the loss of an additional 8 residues), fully digested H2B′ (cleaved after residue 20), H3′, and H4" (which differs from H4′ by the loss of an additional two residues). Trypsin digestion (at points indicated by open arrows [histones designated with a T]) yields fully tailless histones. Trypsin also removes short stretches from the C termini of H2A and H3 (5). (B) SDS-PAGE analysis of histone octamer donor chromatin. Lane U, native histone octamer donor (i.e., H1- and H5-stripped but undigested chromatin); lane 1, histone donor for the tet-no-tails sample obtained by limited clostripain digestion; lane 2, histone donor for the oct-no-tails sample obtained by more-extensive clostripain digestion. H2B′, H2A", and H3′ run together as a single, unresolved broad band (8, 9). Trace amounts of an overdigested product are evident (asterisk). For a similar analysis of the fully tailless histone donor obtained by trypsin digestion, see reference 32. (C) Diagram of the nucleosomal template (see reference 34 for details). Numbers below the drawing represent positions (nucleotide number) of key points in the template; numbers above the drawing represent points along the RNA, including the position of the first U (residue +20) and the runoff product (nt 199). The sequence derives from the 5S RNA gene nucleosome positioning sequence (38) with changes to incorporate a promoter for T7 RNAP (nt 1 to 17) and transcription start site and U-less cassette (nt 18 to 37). The predominant nucleosome position is indicated by the ellipse.