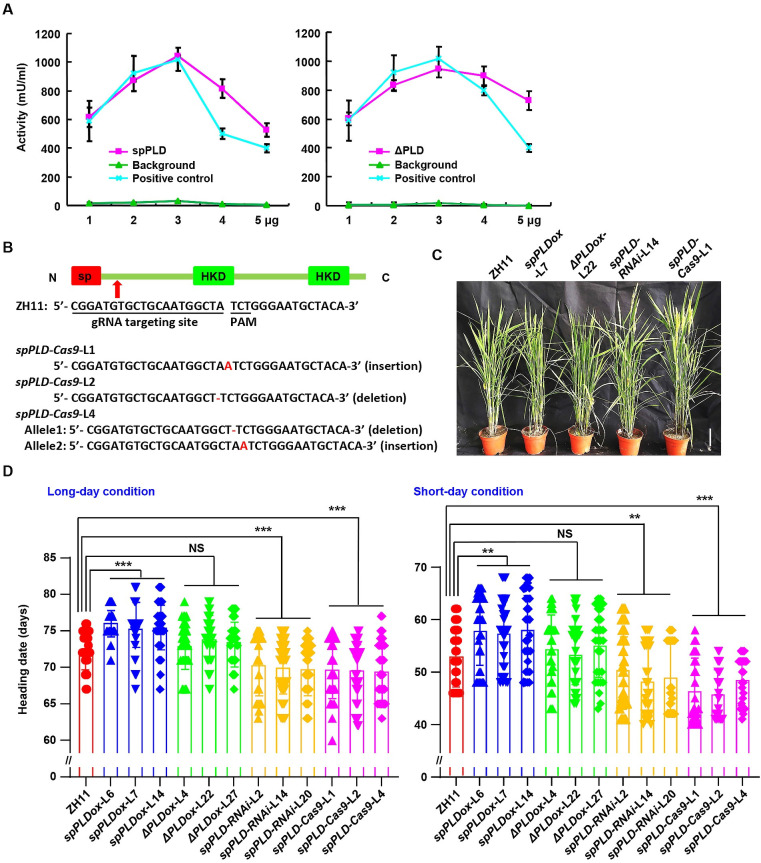
Fig 2. spPLD delays rice heading time.
A. Enzymatic assay showed that both spPLD and ΔPLD (spPLD deleting the signal peptide) present PLD activity. Purified spPLD or ΔPLD (1–5 μg) proteins were used for examination and choline was used as substrate. There is no choline in background (as negative control) and positive control is supplied in assay kit. Experiments were repeated three times and data were shown as mean ± SD (n = 3). B. Sequencing confirmed three mutation lines of spPLD (insertion or deletion of bases), by CRISPR/Cas9. The gRNA targeting site and PAM sequence, and the position of gRNA at spPLD gene, are indicated. C. Phenotypic observation of rice plants with altered spPLD expression under natural long-day condition at heading stage. Representative images were shown. Scale bar = 10 cm. D. Rice plants with altered spPLD expression were grown under natural long-day (left) or short-day (right) conditions and heading date were calculated and statistically analyzed using Tukey’s test (**, p < 0.01; ***, p < 0.001). NS, no significance. Days to flowering were scored when first panicle was bolted, and data were shown as mean ± SD (n = 50).