Fig. 2. Cobamide sharing in model microbial consortia.
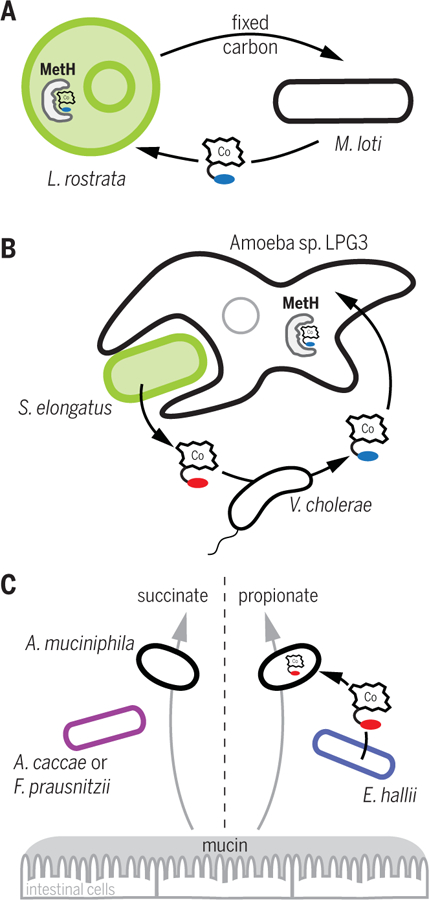
(A) Algae-bacteria mutualism. L. rostrata requires cobalamin produced by M. loti as a cofactor for its methionine synthase (MetH), and M. loti uses fixed carbon produced by L. rostrata (40, 56, 57). (B) Amoeba-prey consortium. V. cholerae remodels pseudocobalamin produced by Synechococcus elongatus to produce cobalamin, which can be used by the amoeba LPG3 for MetH activity (25, 29). (C) Cocultures of A. muciniphila with other human gut bacteria. Cobamide production by E. hallii causes a switch in the mucin degradation product generated by A. muciniphila, from succinate to propionate (62). Cobamide (Co-containing cartoon) with blue indicates cobalamin, and that with red indicates pseudocobalamin.
