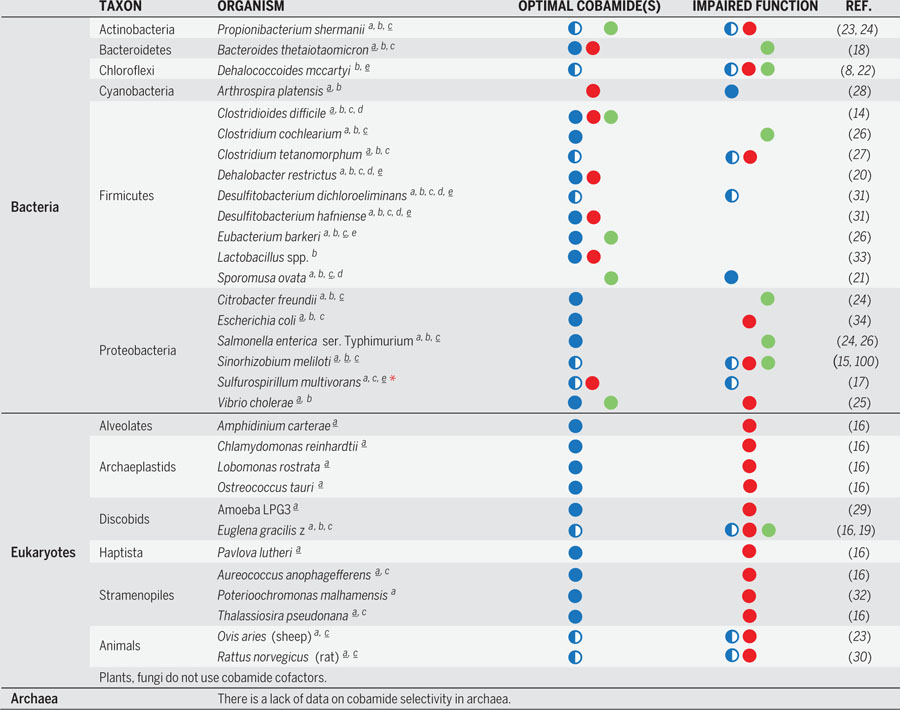
Table 2. Distinct cobamide use by different organisms.
Colored circles correspond to the lower ligand structural classes shown in Fig. 1B (blue, benzimidazolyl; red, purinyl; green, phenolyl cobamides); half-filled circles indicate a subset of the lower ligands within a class (in most cases only one member of each structural class has been tested for function, so the effect of diverse analogs is unknown). Superscripts after species names indicate the cobamide-dependent functions encoded in the genome of each organism, as defined in Table 1 (note, some species have not been sequenced, so they may be annotated incompletely in this table; radical SAM-B12 enzymes indicated by the superscript d are assumed to be involved in secondary metabolite biosynthesis, but not all have been experimentally characterized so some may have other functions). Underlined superscripts indicate the metabolisms that correspond to the selectivity data shown in this table, if known. S. multivorans uses norcobamides, indicated by the red asterisk. Data are from (8, 14–34, 100), as indicated in the rightmost column.
|
