Abstract
Extracellular vesicles (EVs) are capable of transferring cargo from donor to recipient cells, but precisely how cargo content is regulated for export is mostly unknown. For miRNA cargo, we previously showed that when compared to isogenic colorectal cancer (CRC) cells expressing wild-type KRAS, a distinct subset of miRNAs are differentially enriched in EVs from KRAS mutant active CRC cells, with miR-100 being one of the most enriched. The mechanisms that could explain how miR-100 and other miRNAs are differentially exported into EVs have not been fully elucidated. Here, we tested the effect of N6-methyladenosine (m6A) modification on miRNA export into EVs by depletion of METTL3 and ALKBH5, a writer and eraser of m6A modification, respectively. While the effects of ALKBH5 knockdown were quite modest, decreased levels of METTL3 led to reduced cellular and extracellular levels of a subset of miRNAs that contain consensus sequences for m6A modification. Functional testing of EVs prepared from cells expressing shRNAs against METTL3 showed that they were less capable of conferring colony growth in 3D to wild-type KRAS cells and were also largely incapable of conferring the spread of cetuximab resistance. Our data support a role for METTL3 modification on cellular miRNA levels and export of specific miRNAs.
Keywords: miRNA, Base modification, Extracellular vesicle, m6A, RNA, Tumor microenvironment
miRNA, Base modification, Extracellular vesicle, m6A, RNA, Tumor microenvironment
1. Introduction
Recently, miRNAs and other small RNAs have been detected in the extracellular space, usually packaged in extracellular vesicles (EVs), lipoproteins, or protein complexes (Cha et al., 2015b; Crescitelli et al., 2013; Dou et al., 2016; Hinger et al., 2018; Valadi et al., 2007). EVs are a heterogeneous population of nanosized particles secreted from every cell and appear to play a novel role in cell-cell communication by transfer of RNA, lipid, and protein cargo (Maas et al., 2017; Tkach and Thery, 2016). Distinct mechanisms regulating EV cargo content have been proposed (Maas et al., 2017; Shifrin et al., 2013; Simons and Raposo, 2009). We previously showed that EV cargo content is regulated in a KRAS-dependent manner with miR-100 being the most enriched miRNA (Cha et al., 2015b; Demory Beckler et al., 2013; Dou et al., 2016; Hinger et al., 2018). However, we have been unable to identify a specific mechanism that could account for differential miRNA enrichment in EVs. Here, we sought to test whether RNA base modifications might play a role in selective miRNA export.
Over 130 RNA modifications have been identified and recent work has highlighted the importance of understanding the potential role that RNA epitranscriptomics might play in regulating RNA metabolism and gene expression (Barbieri and Kouzarides, 2020; Deng et al., 2018; Holoch and Moazed, 2015; Jonkhout et al., 2017; Nachtergaele and He, 2018; Ries et al., 2019; Roundtree et al., 2017). As with chromatin modifications, there are distinct writers, readers, and erasers of RNA modifications with the best characterized of these involved with N6-methyladenosine (m6A) (Panneerdoss et al., 2018; Roundtree et al., 2017; Zaccara et al., 2019). Methyltransferase-like protein 3 (METTL3) is an RNA methyltransferase that acts as the catalytic subunit of the complex that methylates N6-adenosine (Bokar et al., 1994, 1997; Sibbritt et al., 2013). Alkylation repair homolog protein 5 (ALKBH5) is a demethylase that can reverse m6A in mRNA (Zheng et al., 2013). The majority of work examining the role of m6A modifications has been directed toward its effects on mRNA metabolism, including turnover, splicing, export, and translation (Nachtergaele and He, 2018). For miRNAs, m6A modifications have been found to regulate pri-miRNA processing (Alarcon et al., 2015b; Yuan et al., 2014), but the extent of modifications and any potential roles they might play within mature, processed miRNAs are less clear (Berulava et al., 2015; Konno et al., 2019; Yuan et al., 2014).
We used shRNAs to deplete multiple RNA base modifying enzymes in KRAS mutant CRC cells and found that depletion of METTL3 affected the cellular and extracellular levels of miRNAs containing m6A consensus sequences, including miR-100. We also found that METTL3 knockdown cells were inhibited for growth in 3D and that EVs from knockdown cells were less capable of inducing the spread of cetuximab (CTX) resistance. As the presence of mutant KRAS makes tumors refractory to EGFR blockade by the monoclonal antibody CTX (Lievre et al., 2006), and this excludes CRC patients from CTX treatment (Van Cutsem et al., 2011), the mechanism by which tumor cells act non-autonomously to regulate each other is critical for understanding the evolution of heterogenous tumors in the tumor microenvironment when patients undergo drug treatment. These studies give a mechanistic insight into these issues as the deletion of METTL3 abrogates the ability of these cells to transfer CTX resistance.
2. Results
2.1. miRNA base modifications and EV secretion
To test whether RNA base modifications might play a role in miRNA export, we initially used mass spectrometry and detected the presence of 12 modified ribonucleosides in RNA purified from EVs from mutant (DKO-1) and wild-type (Dks-8) KRAS cells (Figure S1). EVs encompass a variety of particles and for these experiments, we used differential centrifugation to isolate a heterogeneous population of EVs that pelleted after a 17-hour spin at 100,000xg. Because the modifications we detected included abundant tRNA modifications, it was unclear to what extent miRNAs might be modified in those EVs. To better assess whether mature miRNAs might be modified in our EVs, we gel purified 22-23nt RNAs from EV pellets derived from mutant KRAS cells (DKO-1) and performed two-dimensional thin layer chromatography (2D TLC) analysis of modified nucleosides (Figure S2). By radiolabeling RNase T2 products, significant levels of pseudouridine and m6A were identified, in addition to at least five other potential modified nucleosides. Although it is possible that some of the size-selected RNAs might have been derived from non-miRNA sources, our detection of methylated bases in EVs is consistent with reports that mature miRNAs contain m6A (Berulava et al., 2015; Konno et al., 2019). Thus, we decided to test the effects of knockdown of a select group of writers, readers, and erasers of RNA modifications on miRNA export into EVs (Table S1). Except for the EM images in Figure S5, the EVs we analyzed in this paper were derived from pellets via differential centrifugation with a final 17-hour spin at 100,000xg. Nanoparticle tracking analysis was used to quantify particle counts (Figure S3) which showed similar EV secretion across all knockdown lines. The average size of the EVs across all preparations was 133.7 ± 7.9 nm.
2.2. RNA methylation and miRNA export
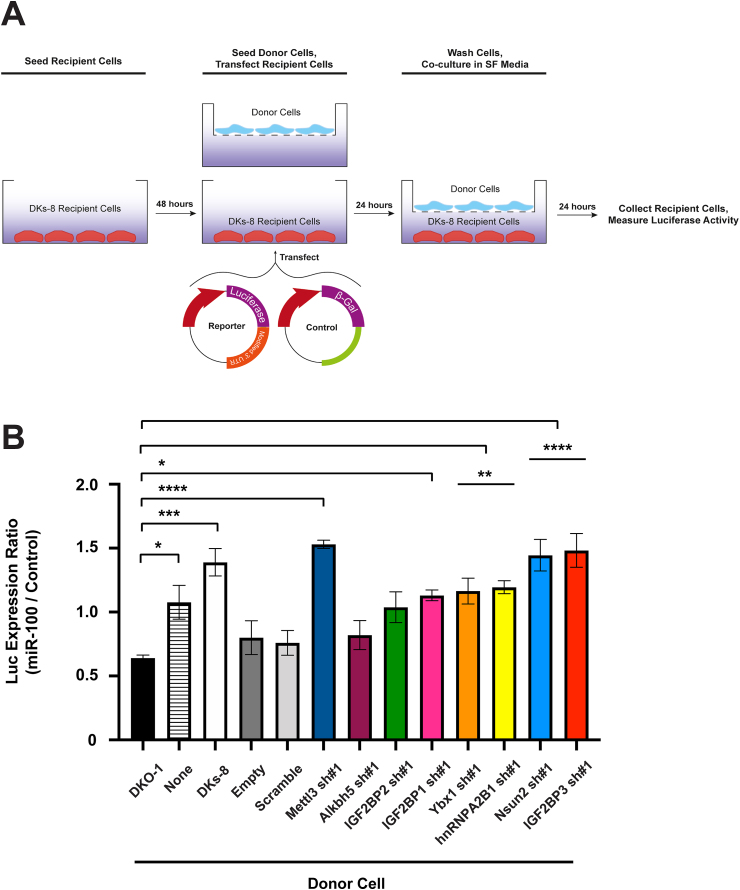
To test whether any of the knockdown cell lines display altered miRNA transfer, we first used a well characterized luciferase reporter in Transwell cultures designed to detect functional transfer of miR-100 from donor KRAS mutant cells to recipient DKs-8 wild-type KRAS cells (Cha et al., 2015b). Recipient cells were transfected with either a luciferase construct containing 3 perfect miR-100 binding sites in the 3′ UTR, or a control construct with a scrambled, non-targeted 3’ UTR sequence lacking any known miRNA binding sites (Figure 1A). Donor knockdown KRAS mutant cells were plated on top of Transwell membranes and luciferase levels were determined in recipient cells after 24 h. We found that 6 of the cell lines showed significantly increased luciferase levels implying inhibition of miR-100 export from donor cells (Figure 1B). A common theme among all 6 of the knockdown lines that showed defects in the miR-100 reporter assay is their role in RNA base methylation or recognition: METTL3 is the catalytic subunit of the N6-methyltransferase complex, the IGF2BP proteins are readers of m6A, hnRNPA2/B1 is thought to be a reader of m6A but may actually bind adjacent to m6A, the Drosophila YBX-1 homolog recognizes 5-methylcytosine, and NSUN2 is a 5-methylcytosine transferase (Alarcon et al., 2015a; Chellamuthu and Gray, 2020; Huang et al., 2018; Kossinova et al., 2017; Shurtleff et al., 2016; Wu et al., 2018; Yuan et al., 2014; Zou et al., 2020) (Table S1).
Figure 1.
Reduced extracellular transfer of miR-100 after knockdown of readers, writers and erasers of RNA modification. Stable shRNA knockdown cell lines were created using mutant KRAS (DKO-1) cells transfected with either an empty shRNA vector, a scrambled control shRNA vector, or vectors encoding shRNAs targeting 14 different proteins (see Table S1). A) Schematic of luciferase reporter assay. B) Luciferase reporter assay. Wild-type KRAS DKs-8 recipient cells were seeded in the bottom of Transwell dishes and co-transfected with vectors expressing β-galactosidase and a luciferase reporter containing either a control 3′UTR or a modified 3′UTR with three perfect miR-100 binding sites. Recipient cells were co-cultured the indicated donor cells for 24 h before cell lysates were collected. Luciferase expression was quantified with decreased expression indicating increased transfer of miR-100. Significance was determined by one-way ANOVA. Data represent mean ± SE, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p = 0.0002, ∗∗∗∗p < 0.0001.
2.3. Knockdown of METTL3 alters levels and extracellular transfer of miR-100
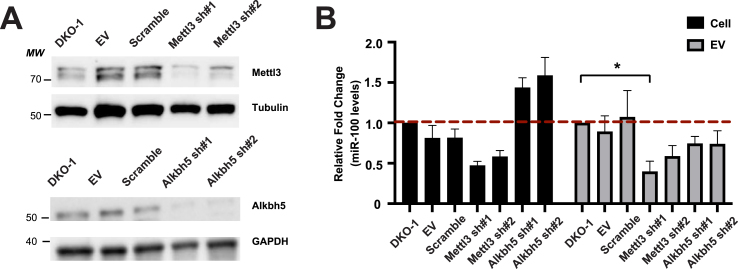
Because 4 of the 6 knockdown lines involved m6A modification and because we detected m6A in 2D TLC analyses (Figure S2), we decided to focus on the effects of decreased METTL3 on miRNA secretion, complemented by similar analyses after knockdown of ALKBH5, an eraser of m6A. The extent of shRNA-mediated knockdown for the two proteins was 60–70% using two independent shRNAs (Figure 2A). Analysis of miR-100 levels by RT/PCR in METTL3 and ALKBH5 knockdown cells and EV pellets showed contrasting differences (Figure 2B). The levels of miR-100 were decreased after knockdown of METTL3 in both cells and EVs, but only rose to significance in EVs from one of the knockdown lines. For ALKBH5, miR-100 levels trended up in cells and down in EVs, but did not reach statistical significance in either case.
Figure 2.
Decreased levels of METTL3 and ALKBH5 alters cellular and EV small RNA levels. A) Western blots were performed on cell lysates from stable lines expressing an empty shRNA vector (Empty), a scrambled control shRNA vector, or two independent shRNAs targeting METTL3 or ALKBH5. Representative immunoblots using antibodies against METTL3, ALKBH5, Tubulin, and GAPDH are shown with the average decrease in METTL3 and ALKBH5 being 60.4 ± 1.1% and 68.4 ± 1.04%, respectively. B) qRT/PCR analysis of miR-100 levels in cellular (black) and EV (gray) samples. Significance was determined by one-way ANOVA. Data represent mean ± SE, n = 3, and ∗p < 0.05.
2.4. Small RNA profiles after knockdown of METTL3 and ALKBH5 are distinct between cells and EVs
To more quantitatively and comprehensively assess the effects of depletion of METTL3 and ALKBH5 on cellular and EV RNA profiles, we conducted RNAseq on libraries created from small RNAs purified from control parental DKO-1 cells and EVs, and from knockdown cells and EVs (Figure S4; Table S2). Analysis of the size distribution of small RNAs purified from control and knockdown cells showed an enrichment for RNAs with a size consistent with miRNA (∼22-23nt), whereas the EV RNA preparations contained peaks corresponding to miRNA, but also included a significant fraction of larger RNAs (30-40nt) corresponding mostly to fragments derived from tRNA and rRNA (Figure S4A). Total read numbers indicated sufficient coverage with over 5 million reads for almost every sample (Figure S4B; Table S2). When quantifying read numbers across the main categories of small RNAs found in the respective libraries, the percentage composition for the different classes of RNA was similar between both the control and knockdown cells and between the control and knockdown EVs (Figure S4C). However, there was a clear difference in the read distribution comparing cellular small RNAs to EV small RNAs, consistent with Principal Component (PC) analysis which showed distinct differences in RNA profiles comparing cells to EVs (Figure S4D). The cellular profiles tended to cluster together whereas the EV profiles showed greater variation by PC analysis.
2.5. METTL3 knockdown causes downregulation of miRNAs containing m6A consensus sites
We next analyzed the small RNAseq data to identify those miRNAs that were most upregulated or downregulated in cells or EVs after knockdown of either METTL3 or ALKBH5. After applying minimal expression threshold levels and normalizing for differential expression using DESeq2, the effects of ALKBH5 knockdown were quite modest with no miRNAs showing a log2 fold change greater than +1 or -1 in cells and only one miRNA (miR-3182) showing a log2 fold change greater than +1 in EVs. When we performed the same analysis on small RNAseq data after METTL3 knockdown, no miRNAs were significantly upregulated greater than log2 fold change +1 in either cells or EVs. In contrast, a distinct subset of miRNAs were significantly downregulated (log2 fold change > -1) in both cells and EVs after METTL33 knockdown (Table 1). Remarkably, 5 of the top 7 most downregulated miRNAs in EVs contain consensus sequences for m6A modification with a 6th miRNA (let-7e-5p) having an m6A consensus sequence in the immediate downstream loop sequence (pre-let-7e-5p) (Table 1). In cells, 10 miRNAs were downregulated with a log2 fold change greater than -1. Five of those miRNAs contain m6A consensus sites in the mature miRNA with 3 containing m6A sites in the immediate downstream precursor loop sequence. Given that we began these experiments attempting to understand differential export of miR-100 in KRAS mutant cells, it is striking that knockdown of METTL3 had a significant effect on miR-100. However, because both cellular and EV levels were affected, the data are not conclusive for a direct role for m6A modification in overall miRNA export, but suggest that m6A modification plays a role in cellular processing, stability, and/or export of these miRNAs.
Table 1.
Downregulated miRNAs contain m6A consensus sequences.
| Cells | |||||
|---|---|---|---|---|---|
| miRNA | Read Base Mean | Sequence | FC | P-value | Adjusted P-value |
| miR-125b-5p | 360.877 | UCCCUGAGACCCUAACUUGUGA | -1.7 | 5.07E-09 | 2.91E-06 |
| miR-100-5p | 1252.25 | AACCCGUAGAUCCGAACUUGUG | -1.7 | 3.7E-08 | 1.06E-05 |
| let-7e-5p | 410.781 | UGAGGUAGGAGGUUGUAUAGUUgaggaggaca | -1.3 | 1.91E-06 | 0.00036 |
| miR-4521 | 114.646 | GCUAAGGAAGUCCUGUGCUCAGuuuuguagcaucaaaacu | -1.0 | 6.32E-05 | 0.008 |
| miR-142-5p | 121.025 | CAUAAAGUAGAAAGCACUACU | -1.2 | 7.13E-05 | 0.008 |
| miR-146a-5p | 149.772 | UGAGAACUGAAUUCCAUGGGUU | -1.0 | 9.86E-05 | 0.009 |
| miR-99b-5p | 844.702 | CACCCGUAGAACCGACCUUGCG | -1.2 | 0.00017868 | 0.015 |
| miR-125a-5p | 1040.74 | UCCCUGAGACCCUUUAACCUGUGA | -1.0 | 0.00045275 | 0.026 |
| miR-135b-5p | 127.410 | UAUGGCUUUUCAUUCCUAUGUGAuugcugucccaaacu | -1.1 | 0.00238943 | 0.099 |
|
miR-32-5p |
241.298 |
UAUUGCACAUUACUAAGUUGCA |
-1.0 |
0.0222186 |
0.308 |
| EVs | |||||
| miRNA |
Expression Level |
Sequence |
FC |
P-value |
Adjusted P-value |
| miR-100-5p | 1252.25 | AACCCGUAGAUCCGAACUUGUG | -1.5 | 1.09E-06 | 0.00074 |
| miR-99b-5p | 844.702 | CACCCGUAGAACCGACCUUGCG | -1.4 | 1.07E-05 | 0.003 |
| miR-193b-3p | 582.185 | AACUGGCCCUCAAAGUCCCGCU | -1.0 | 1.39E-05 | 0.003 |
| miR-125b-5p | 360.877 | UCCCUGAGACCCUAACUUGUGA | -1.2 | 7.31E-05 | 0.001 |
| let-7e-5p | 410.781 | UGAGGUAGGAGGUUGUAUAGUUgaggaggaca | -1.1 | 0.00014876 | 0.011 |
| miR-125a-5p | 1040.74 | UCCCUGAGACCCUUUAACCUGUGA | -1.2 | 0.00020023 | 0.013 |
| miR-224-5p | 1469.11 | UCAAGUCACUAGUGGUUCCGUUUAG | -1.0 | 0.00274813 | 0.098 |
Downregulated miRNAs in METTL3 knockdown cells and EVs compared to DKO-1 cells and EVs. Expression levels (Read Base Mean) are the average number of RNAseq reads from 3 independent libraries. The sequence of downregulated miRNAs is shown along with the log2 fold change (FC), p-values, and adjusted p-values when comparing cell and EV levels between METTL3 knockdown cells and control DKO-1 cells after DESeq2 normalization. Highlighted in red are the m6A consensus sequences present in the mature miRNA sequence (upper case) or the immediately adjacent loop sequence in the pre-miRNA.
2.6. Knockdown of METTL3 reduces EV-mediated anchorage-independent growth
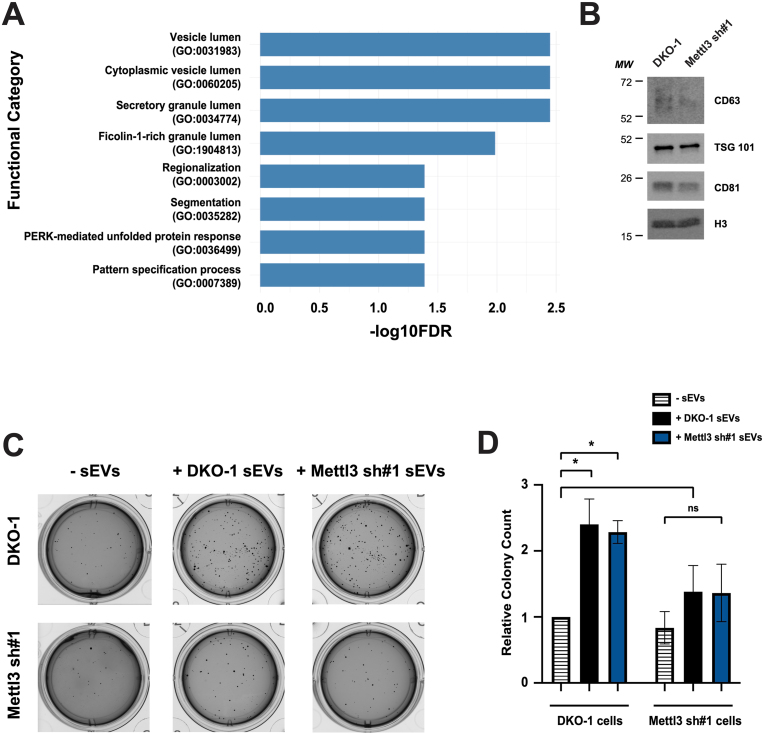
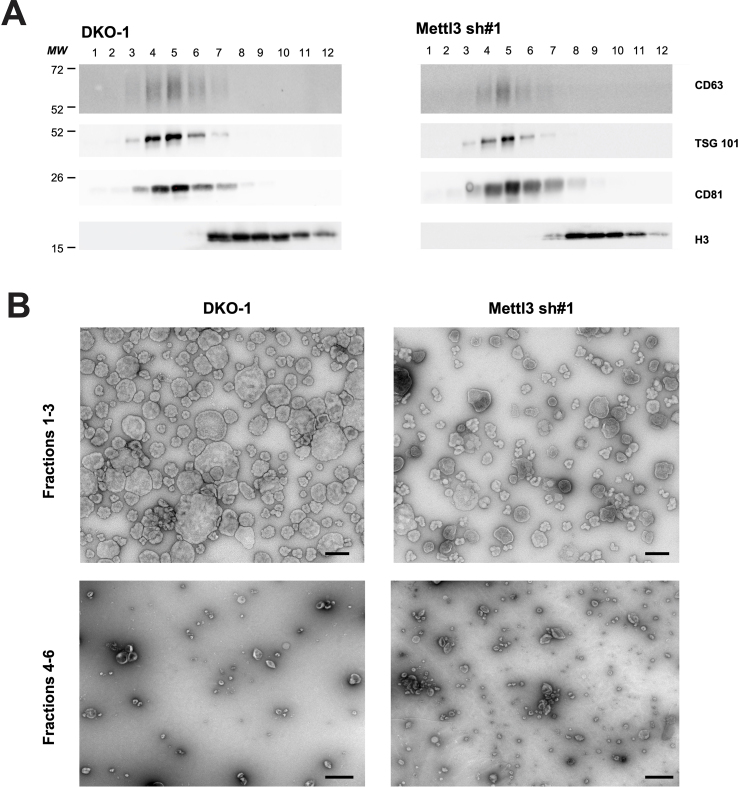
The observed changes in cellular small RNAs prompted us to perform mRNAseq on our knockdown cells to identify gene expression changes (Figure 3, Table S3). We used the WebGestalt functional enrichment analysis tool (Liao et al., 2019) to identify Gene Ontology categories that were the most downregulated (Figure 3A). The most affected pathways included four secretory cascade categories. Because the cellular genes most affected involved the secretory cascade, we decided to determine whether changes due to METTL3 knockdown would alter cellular behavior and EV tumor microenvironment interactions mediated by EVs. Previously, we showed that EVs from mutant KRAS DKO-1 cells can promote proliferation and anchorage-independent growth (Demory Beckler et al., 2013; Higginbotham et al., 2011) and that Rab13 regulation of EV secretion can inhibit cell growth in soft agar (Hinger et al., 2020). Further, analysis of the Cancer Genome Atlas (TCGA) database showed that METTL3 expression is increased in metastatic colorectal cancer and is associated with a poor prognosis (Li et al., 2019). Thus, we compared the growth of METTL3 knockdown cells to control DKO-1 cells in soft agar and also compared the growth of these cells in 3D after exposure to EVs from both cells. For this, we purified and characterized EVs from DKO-1 and METTL3 knockdown cells using western blots with antibodies against classical EV (exosome) markers (Figure 3B). By both western blot analysis and morphology using electron microscopy, no significant differences were observed between EVs from the control and knockdown cells (Figure 3B; Figure S5).
Figure 3.
Knockdown of METTL3 reduces EV-mediated anchorage-independent growth. A) Gene ontology analysis of significant biological processes affected in METTL3 knockdown cells compared to DKO-1 control cells after total RNAseq. B) Immunoblots of EVs from DKO-1 or METTL3 knockdown cells using antibodies against CD63, TSG101, CD81, and H3. C,D) DKO-1 and METTL3 knockdown cells (500 cells/well) were grown in soft agar for 2 weeks in the presence or absence of EVs derived from either DKO-1 cells or METTL3 knockdown cells. Representative images (C) and quantification of colony counts (D) are shown. Significance was determined by one-way ANOVA. Equivalent amounts of EVs were added. Data represent mean ± SE, n = 3, ∗p < 0.05.
Consistent with previous results, DKO-1 cells were capable of growth in soft agar and colony numbers increased after exposure to self EVs (Figure 3C,D). Increased growth of DKO-1 cells was also observed after exposure to an equivalent amount of EVs from METTL3 knockdown cells (Figure 3C,D). In contrast, METTL3 knockdown cells did not grow as well in soft agar and the effect of exposure to EVs from either DKO-1 or METTL3 knockdown cells did not significantly increase growth (Figure 3C,D). These data support the conclusion that depletion of METTL3 causes a cell-autonomous decrease in the ability to grow in soft agar compared to control DKO-1 cells, even when exposed to DKO-1 control EVs. These results also suggest that METTL3 knockdown causes a decreased ability to receive soft agar-dependent, growth inducing signals from DKO-1 EVs.
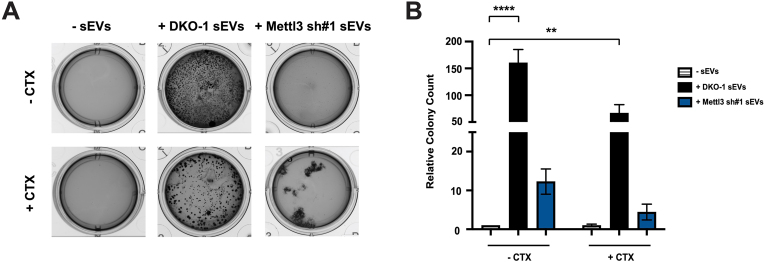
2.7. Knockdown of METTL3 reduces EV-mediated transfer of cetuximab resistance
Since two of the top downregulated miRNAs in EVs after METTL3 knockdown are miR-100 and miR-125b, we next decided to test the effects of EVs from DKO-1 and METTL3 on anchorage-independent growth of wild-type KRAS cells (DKs-8) and on the transfer of cetuximab resistance. DKs-8 cells form minimal, if any, colonies in soft agar unless exposed to EVs from mutant KRAS DKO-1 cells which contain high levels of miR-100 and miR-125b (Cha et al., 2015a; Demory Beckler et al., 2013). Further, growth of DKs-8 cells is susceptible to cetuximab, a monoclonal antibody against the Epidermal Growth Factor Receptor (EGFR). We previously showed that upregulation of miR-100 and miR-125b can induce resistance to cetuximab (Lu et al., 2017). When DKs-8 cells were exposed to EVs from DKO-1 cells, they formed colonies in soft agar and were resistant to cetuximab (Figure 4A,B). In contrast, exposure to EVs from METTL3 knockdown cells did not stimulate growth in soft agar or confer resistance to cetuximab (Figure 4A,B). Interestingly, we occasionally detected a few surviving colonies in the presence of cetuximab after exposure to EVs from METTL3 knockdown cells but they consistently displayed an unusual morphology (Figure 4A). These data support the conclusion that the cargo contained within EVs isolated from METTL3 knockdown cells is not able to drive colony growth in soft agar and is not able to confer drug resistance on recipient cells, having lost components present in DKO-1 control EVs that are able to provide these functions. The decreased levels of miR-100 and miR-125b are consistent with their action in regulating these EV functions.
Figure 4.
Knockdown of METTL3 reduces EV-mediated transfer of cetuximab resistance. Wild-type KRAS DKs-8 cells (∼2000 cells/well) were grown in soft agar for 2 weeks in the presence or absence of EVs derived from either control DKO-1 cells or METTL3 knockdown cells. Representative images (A) and quantification of colony counts (B) are shown. Significance was determined by one-way ANOVA. Equivalent amounts of EVs were added. Data represent mean ± SE, n = 3, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
3. Discussion
Numerous studies have demonstrated consistent differential enrichment of specific miRNAs within EVs (Cha et al., 2015b; Kosaka et al., 2010; McKenzie et al., 2016; Santangelo et al., 2016; Shurtleff et al., 2016; Skog et al., 2008; Squadrito et al., 2014; Valadi et al., 2007; Wei et al., 2017). While such changes could be due to differential miRNA stability and/or some means of passive differential loading of miRNAs into EVs, it remains a key focus of current research to test potential mechanisms that specifically regulate miRNA export (Mateescu et al., 2017). RNA sequence motifs, roles for specific RNA binding proteins, and differential mRNA target expression have all been proposed to drive selective miRNA export, but no universal mechanism has been identified (Batagov et al., 2011; Bolukbasi et al., 2012; Kossinova et al., 2017; Santangelo et al., 2016; Shurtleff et al., 2016, 2017; Squadrito et al., 2014; Statello et al., 2018; Villarroya-Beltri et al., 2013). This might mean that all export control is context-specific or it might mean that we have thus far failed to recognize mechanistic similarities. For our CRC model, we detected KRAS-dependent differential miRNA enrichment, but we were unable to identify a unique RNA sequence motif or RNA binding protein that could mediate selective export (Cha et al., 2015a). This prompted us to test whether RNA base modifications or the combination of base modifications and recognition by specific RNA binding proteins might drive deposition into EVs. While we show that depletion of METTL3 leads to downregulation of miRNAs that contain m6A consensus sites, many of those same miRNAs are downregulated in cells. Thus, even though there might be a direct regulatory role for base modifications in miRNA export, it is not possible to make a firm conclusion with the given data. Disentangling direct or indirect roles for m6A modification between cellular and EV RNA could potentially be performed with differential isotopic labeling or other methods, but are beyond the current scope of this work. Similarly, it will be difficult to utilize site-directed mutagenesis to alter specific adenosine residues because single nucleotide changes could alter normal miRNA function by affecting base pairing with targets, altering the binding of specific RNA binding proteins, altering secondary structure, stability, or some combination thereof (Harcourt et al., 2017; Liu et al., 2015). Despite limitations on conclusions related to miRNA export, our data are nevertheless consistent with a role for m6A modification in the stability of mature miRNAs (Berulava et al., 2015; Konno et al., 2019), in addition to supporting a role for m6A modification during miRNA biogenesis (Alarcon et al., 2015b; Yuan et al., 2014).
m6A modifications are abundant across many different types of RNA (Nachtergaele and He, 2018; Zaccara et al., 2019). Readers of m6A have been proposed to affect the stability of mRNAs (Heck et al., 2020; Huang et al., 2018; Ke et al., 2017; Mauer et al., 2017; Wang et al., 2014; Zhao et al., 2017), but might also serve as nucleation sites for the formation of condensates that might underlie non-membranous compartments or complexes such as P-bodies, stress granules, or nuclear speckles that can play a role in RNA stability, including miRNA half-life (Ries et al., 2019). The presence of readers, writers and erasers of m6A suggest the potential for dynamic changes in gene expression at the RNA level (Meyer and Jaffrey, 2014; Roundtree et al., 2017; Zhou et al., 2015). Related to our work on CRC, dynamic changes in m6A modification have been proposed to regulate cancer growth and progression and serve as potential anti-cancer drug targets (Barbieri and Kouzarides, 2020; Boriack-Sjodin et al., 2018; Deng et al., 2018; Panneerdoss et al., 2018).
EVs are now thought to represent a heretofore unappreciated form of cell-cell communication, both locally within the tumor microenvironment and the immune synapse, and at a distance in the metastatic niche (Gutierrez-Vazquez et al., 2013; Maas et al., 2017; Maia et al., 2018; McAllister and Weinberg, 2014; Shurtleff et al., 2018; Wortzel et al., 2019). We previously showed that exposure of wild-type KRAS cells to EVs from mutant KRAS cells could induce growth and proliferation in wild-type KRAS cells (Demory Beckler et al., 2013; Higginbotham et al., 2011). In a model of cetuximab resistance, we showed that dramatic upregulation of cellular levels of miR-100 and miR-125b caused activation of Wnt signaling and drug resistance (Lu et al., 2017). Here, we show that decreased levels of METTL3 resulted in the secretion of EVs that were not as capable of promoting anchorage-independent growth and also nearly abolished transfer of cetuximab resistance. The fact that miR-100 and miR-125b were two of the top downregulated miRNAs after METTL3 knockdown support the finding that transfer of miR-100 and miR-125b could be, at least in part, responsible for transfer of cetuximab resistance, although protein cargo may also contribute to resistance (Lu et al., 2017).
We have found that knockdown of METTL3 affects the abundance of many classes of RNA, both cellular and extracellular. This is consistent with widespread m6A modification of RNA and the fact that CRISPR/Cas9 mediated knockout of METTL3 in our CRC lines is lethal. Knockdown of METTL3 affects miRNA levels in cells and EVs with the most affected being those that contain m6A consensus sites, including miR-100 and miR-125b which are two of the most enriched miRNAs in EVs in CRC cells. Future experiments will be devoted to identify specific adenosine residues that are modified in these miRNAs and the development of technology to directly determine whether m6A modification of these bases is required for export.
Declarations
Author contribution statement
Jessica J. Abner: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jeffrey L. Franklin, Alissa M. Weaver, James G. Patton: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Margaret A. Clement, Ryan M. Allen: Performed the experiments; Analyzed and interpreted the data.
Scott A. Hinger: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Xiao Liu: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Stefanie Kellner, Junzhou Wu: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
John Karijolich: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Qi Liu: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Kasey C. Vickers: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Peter Dedon, Robert J. Coffey: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by grants from the National Institutes of Health and the National Cancer Institute including P01 CA229123 to Drs. Alissa M. Weaver, Robert J. Coffey and James G. Patton and by P50 CA236733, R35 CA197570, and UG3 CA241685 to Robert J. Coffey and R01 ES031529 and R01 ES026856 to Peter C. Dedon. Mass spectrometric analyses were performed in the MIT Center for Environmental Health Sciences, which is supported by NIEHS grant P30 ES002109.
Data availability statement
The RNAseq datasets generated during this study are available at GEO (Accession numbers: GSE166643 and GSE182382).
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Dr. Evan Krystofiak of the Cell Imaging Shared Resource core for assistance with processing TEM images and the Weaver, Patton, and Coffey labs for discussion, suggestions, and comments.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Fig S5.
References
- Alarcon C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I., Kouzarides T. Role of RNA modifications in cancer. Nat. Rev. Cancer. 2020;20:303–322. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- Batagov A.O., Kuznetsov V.A., Kurochkin I.V. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genom. 2011;12(Suppl 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava T., Rahmann S., Rademacher K., Klein-Hitpass L., Horsthemke B. N6-adenosine methylation in MiRNAs. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Bolukbasi M.F., Mizrak A., Ozdener G.B., Madlener S., Ströbel T., Erkan E.P., Fan J.-B., Breakefield X.O., Saydam O. miR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids. 2012;1:e10. doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin P.A., Ribich S., Copeland R.A. RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug Discov. 2018;17:435–453. doi: 10.1038/nrd.2018.71. [DOI] [PubMed] [Google Scholar]
- Cha D.J., Franklin J.L., Dou Y., Liu Q., Higginbotham J.N., Beckler M.D., Weaver A.M., Vickers K., Prasad N., Levy S., et al. KRAS-dependent sorting of miRNA to exosomes. eLife. 2015;4 doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha D.J., Franklin J.L., Dou Y., Liu Q., Higginbotham J.N., Demory Beckler M., Weaver A.M., Vickers K., Prasad N., Levy S., et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4 doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellamuthu A., Gray S.G. The RNA methyltransferase NSUN2 and its potential roles in cancer. Cells. 2020;9:1758. doi: 10.3390/cells9081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R., Lasser C., Szabo T.G., Kittel A., Eldh M., Dianzani I., Buzas E.I., Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles. 2013;2:20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M., Higginbotham J.N., Franklin J.L., Ham A.J., Halvey P.J., Imasuen I.E., Whitwell C., Li M., Liebler D.C., Coffey R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Su R., Weng H., Huang H., Li Z., Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M., Prasad N., Levy S., Coffey R.J., Patton J.G., et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Vazquez C., Villarroya-Beltri C., Mittelbrunn M., Sanchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 2013;251:125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt E.M., Kietrys A.M., Kool E.T. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A.M., Russo J., Wilusz J., Nishimura E.O., Wilusz C.J. YTHDF2 destabilizes m(6)A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells. RNA. 2020;26:739–755. doi: 10.1261/rna.073502.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham J.N., Demory Beckler M., Gephart J.D., Franklin J.L., Bogatcheva G., Kremers G.J., Piston D.W., Ayers G.D., McConnell R.E., Tyska M.J., et al. Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 2011;21:779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger S.A., Abner J.J., Franklin J.L., Jeppesen D.K., Coffey R.J., Patton J.G. Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Sci. Rep. 2020;10:15804. doi: 10.1038/s41598-020-72503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger S.A., Cha D.J., Franklin J.L., Higginbotham J.N., Dou Y., Ping J., Shu L., Prasad N., Levy S., Zhang B., et al. Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Rep. 2018;25:715–725 e714. doi: 10.1016/j.celrep.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M. The RNA modification landscape in human disease. RNA. 2017;23:1754–1769. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Pandya-Jones A., Saito Y., Fak J.J., Vagbo C.B., Geula S., Hanna J.H., Black D.L., Darnell J.E., Jr., Darnell R.B. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno M., Koseki J., Asai A., Yamagata A., Shimamura T., Motooka D., Okuzaki D., Kawamoto K., Mizushima T., Eguchi H., et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019;10:3888. doi: 10.1038/s41467-019-11826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossinova O.A., Gopanenko A.V., Tamkovich S.N., Krasheninina O.A., Tupikin A.E., Kiseleva E., Yanshina D.D., Malygin A.A., Ven'yaminova A.G., Kabilov M.R., et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta. 2017;1865:664–673. doi: 10.1016/j.bbapap.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N., Chen Z.H., Zeng Z.L., Wang F., Zheng J., et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievre A., Bachet J.B., Le Corre D., Boige V., Landi B., Emile J.F., Cote J.F., Tomasic G., Penna C., Ducreux M., et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhao X., Liu Q., Li C., Graves-Deal R., Cao Z., Singh B., Franklin J.L., Wang J., Hu H., Yang M., et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate Cetuximab resistance via Wnt/beta catenin signaling. Nat. Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S.L.N., Breakefield X.O., Weaver A.M. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J., Caja S., Strano Moraes M.C., Couto N., Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front. Cell Dev. Biol. 2018;6:18. doi: 10.3389/fcell.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Kowal E.J., van Balkom B.W., Bartel S., Bhattacharyya S.N., Buzas E.I., Buck A.H., de Candia P., Chow F.W., Das S., et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., et al. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister S.S., Weinberg R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie A.J., Hoshino D., Cha D.J., Franklin J.L., Coffey R.J., Patton J.G., Weaver A.M. KRAS-MEK signaling controls Ago2 and miRNA sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., He C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss S., Eedunuri V.K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., Abdelfattah N., Onyeagucha B.C., Cui X., Lai Z., et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries R.J., Zaccara S., Klein P., Olarerin-George A., Namkoong S., Pickering B.F., Patil D.P., Kwak H., Lee J.H., Jaffrey S.R. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Shifrin D.A., Jr., Demory Beckler M., Coffey R.J., Tyska M.J. Extracellular vesicles: communication, coercion, and conditioning. Mol. Biol. Cell. 2013;24:1253–1259. doi: 10.1091/mbc.E12-08-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5 doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M.J., Temoche-Diaz M.M., Schekman R. Extracellular vesicles and cancer: caveat lector. Annu. Rev. Cell Biol. 2018;2:395–411. [Google Scholar]
- Shurtleff M.J., Yao J., Qin Y., Nottingham R.M., Temoche-Diaz M.M., Schekman R., Lambowitz A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbritt T., Patel H.R., Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip. Rev. RNA. 2013;4:397–422. doi: 10.1002/wrna.1166. [DOI] [PubMed] [Google Scholar]
- Simons M., Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R., Ibberson M., De Palma M. Endogenous RNAs modulate MicroRNA sorting to exosomes and transfer to Acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- Statello L., Maugeri M., Garre E., Nawaz M., Wahlgren J., Papadimitriou A., Lundqvist C., Lindfors L., Collen A., Sunnerhagen P., et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M., Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E., Kohne C.H., Lang I., Folprecht G., Nowacki M.P., Cascinu S., Shchepotin I., Maurel J., Cunningham D., Tejpar S., et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J. Clin. Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.Y., Batagov A.O., Schinelli S., Wang J.T., Wang Y., El Fatimy R., Rabinovsky R., Balaj L., Chen C.C., Hochberg F., et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017;8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Tang H., Xing J., Fan X., Cai X., Li Q., Han P., Luo Y., Zhang Z., Jiang B., et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol. Cell Biol. 2014;34:3630–3641. doi: 10.1128/MCB.00243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Tu R., Duan B., Yang Z., Ping Z., Song X., Chen S., Price A., Li H., Scott A., et al. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc. Natl. Acad. Sci. U. S. A. 2020;117:3603–3609. doi: 10.1073/pnas.1910862117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq datasets generated during this study are available at GEO (Accession numbers: GSE166643 and GSE182382).