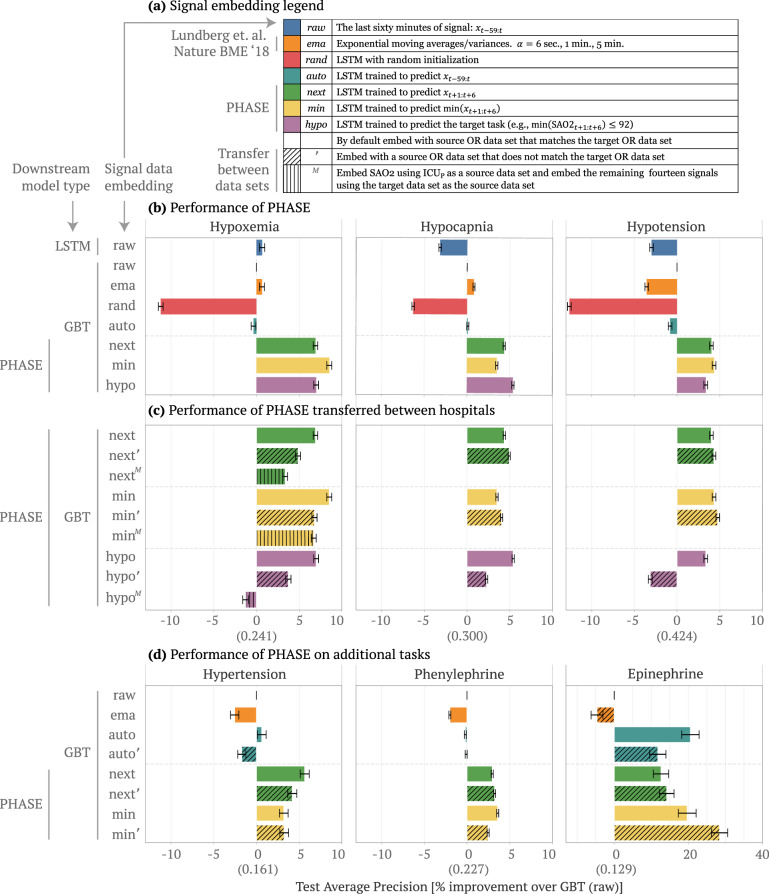
Fig. 2. Performance of PHASE embedding models.
Comparing the performance of downstream models trained with different embeddings of physiological signals concatenated to static features. We report the average precision (% improvement over GBT model trained with raw signal data, 99% confidence intervals from bootstrapping the test set). We use OR0 and OR1 as target datasets and then aggregate across both by averaging the resultant means and standard errors of the % improvement. a The upstream embedding models we use to extract the physiological signal features where raw is the identity function, ema is an exponential moving average, and the rest are LSTMs trained in specific ways.b The performance of downstream prediction models for a variety of standard embedding approaches (when the source hospital is the same as the target hospital). We compare combinations of downstream models and embeddings for three adverse surgical outcomes (hypoxemia, hypocapnia, and hypotension). c The performance of transferred embedding (next', nextM, min', minM, hypo', and hypoM) vs. non-transferred (next, min, and hypo) models for the above three adverse outcomes. In the transferred approaches the source hospital is different to the target hospital. d Performance of approaches for standard and transferred embedding on additional outcomes: hypertension (high, rather than low, blood pressure); phenylephrine and epinephrine (doctor action prediction). We do not evaluate hypo embeddings in this setting, because the outcomes are not “hypo” events. Model architectures in Supplementary Note 6. We report the average precision value of the raw model in parenthesis on the x-axis.