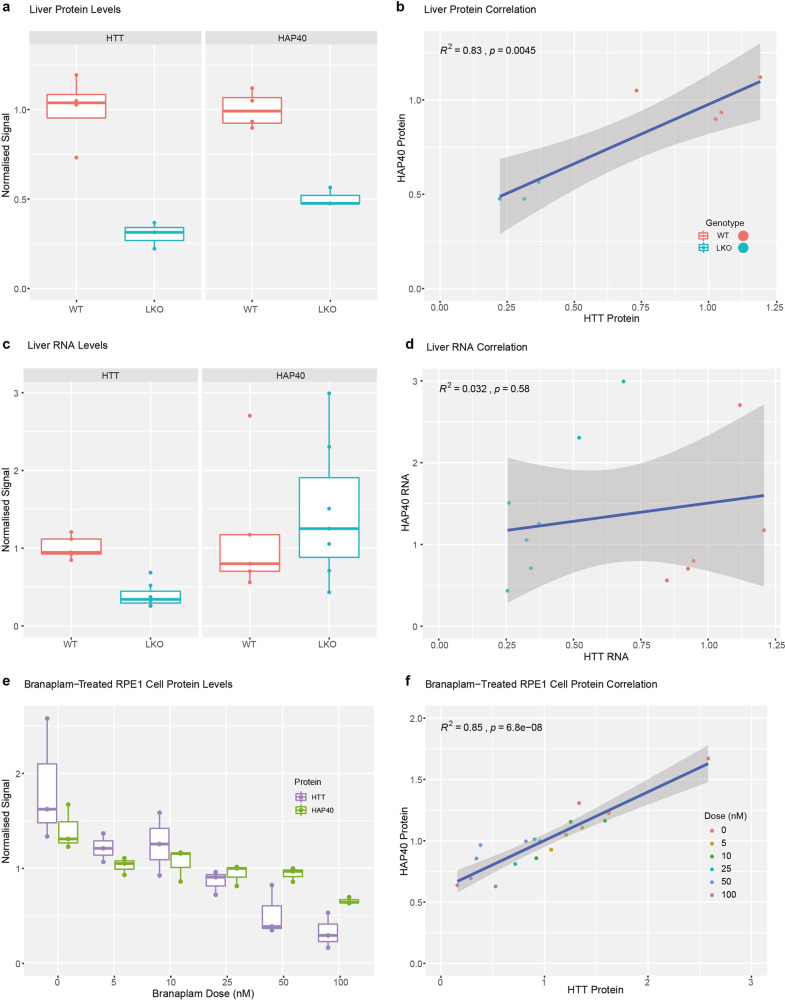
Fig. 1. HTT lowering reduces HAP40 protein levels but not mRNA levels in vivo.
a Htt and Hap40 protein levels in the liver of WT and hepatocyte-specific Htt knockout (LKO) mouse. Hepatocytes make up roughly 80% of liver mass82, and LKO liver tissue shows ~70% reduction in HTT levels while HAP40 is reduced ~50%. The reduction in both instances is statistically significant (unpaired t test; t(4.0407) = 6.6184, p = 0.002607 and t(4.5721) = 8.3375, p = 0.0006203, respectively). b HTT lowering drives reduction of HAP40 protein (R2 = 0.83, F(1,5) = 24.05, p = 0.0045). c Liver Htt and Hap40 RNA levels assessed by quantitative PCR. Htt RNA levels are reduced ~60% in LKO mice (unpaired t test; t(9.039) = 6.9096, p = 6.845 × 10−05), while Hap40 RNA levels remain unchanged. d Lowering Htt RNA levels does not drive Hap40 RNA reduction (R2 = 0.03, F(1,10) = 0.347, p = 0.5757). **p < 0.01, ***p < 0.001. e Western blot quantified HTT and HAP40 protein levels in human RPE1 cells after 72-h treatment with the HTT-lowering drug branaplam or DMSO control (0 nM). f Data points from e plotted showing HTT lowering by branaplam drives reduction of HAP40 protein (R2 = 0.85, F(1,16) = 87,67, p = 6.8 × 10−08). For a, c, e, boxplot hinges correspond to the first and third quartiles of the data, with the horizontal line indicating the median. Whiskers correspond to the smallest and largest datapoint within 1.5 times the interquartile range from the corresponding hinge. Datapoints laying outside the whiskers are >1.5 times the interquartile range from the corresponding hinge.