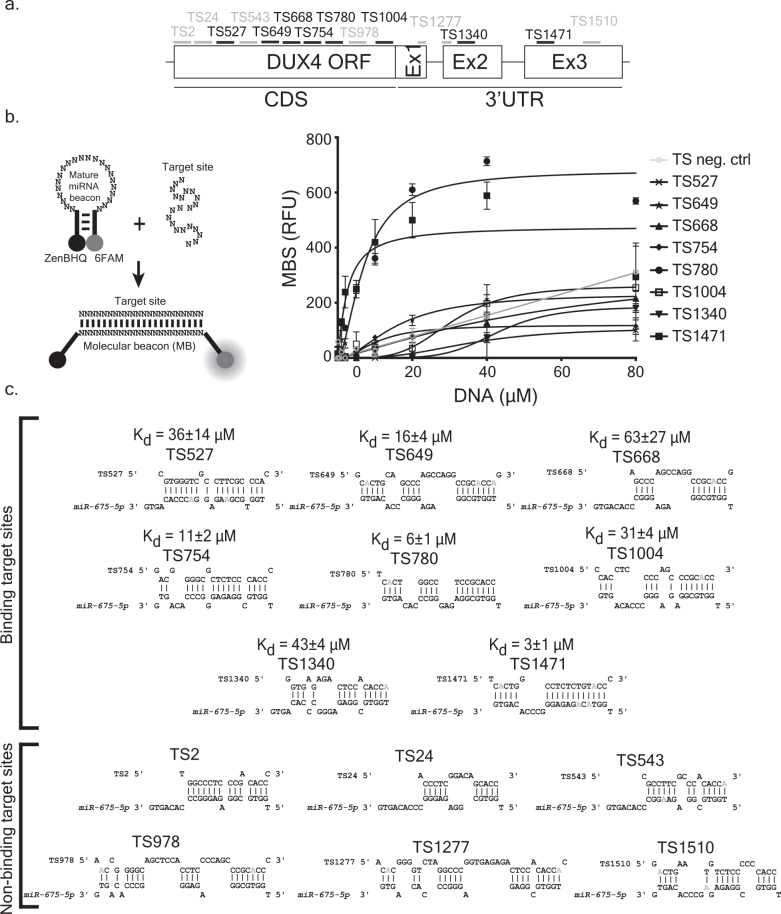
Fig. 2. miR-675-5p targets multiple binding sites in the DUX4 ORF and 3′UTR.
a Schematic of DUX4 sequence showing predicted target site (TS) positions for miR-675-5p. b (Left) Molecular beacon binding assay used to determine miR-675-5p binding to DUX4. Unbound, the molecular beacon folds into a stem loop structure that brings the ZenBHQ quencher in close proximity to a the 6FAM fluorophore, thereby preventing 6FAM fluorescence. We incorporated the mature sequence of miR-675-5p in the MB loop sequence. Hybridization of the MB to a complementary DUX4 target sequence separates the fluorophore and quencher, allowing fluorescence emission, which is quantified as a measure of binding. (Right) The graph shows binding of the mature miR-675-5p molecular beacon to target sites shown in (a). miR-675-5p bound 8 DUX4 target sites (6 in the ORF and 2 in the 3′UTR). Six predicted TS did not bind to miR-675-5p (Supplementary Fig. 3b and Supplementary Data 1 for TS position and sequence). The TS neg. ctrl is a random sequence. Curves are fit into one site-specific binding with Hill slope equation. Each data point represents mean ± SD (N = 3 independent experiments). c Base-pairing of miR-675-5p and predicted DUX4 target sites used in the molecular beacon assay. The binding affinity (Kd) of miR-675-5p molecular beacon to each target site was determined by subtracting background fluorescent signal from the molecular beacon signal (MBS), expressed in relative fluorescent units (RFU). The Kd corresponds to the TS concentration (μM) required to reach half of maximum fluorescence. We generated RNA “mimic” bases in the miR-675-5p:TS pair and replaced “G” nucleotides with “A” nucleotides (in gray) whenever the “G” is facing a “T”29. Source data are provided as a Source data file.