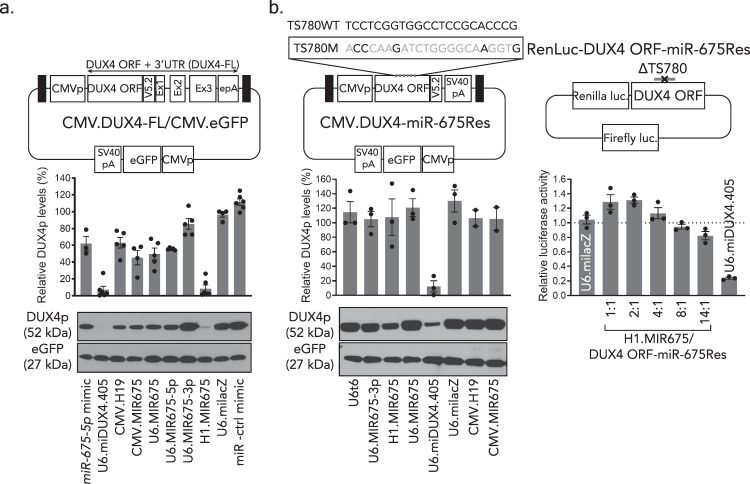
Fig. 3. miR-675-5p specifically targets DUX4 and reduces its protein and mRNA levels in vitro.
a, b Blinded western blots detecting V5 epitope-tagged DUX4 and an eGFP transfection control in the presence of miR-675 or control constructs. a Proteins were harvested from HEK293 cells 48 h after co-transfection with the CMV.DUX4-FL/CMV.eGFP expression plasmid and indicated miR-675 or control plasmids. Black boxes, AAV2 inverted terminal repeats (ITR). CMVp, cytomegalovirus promoter. Western blot membranes were stained with antibodies to the V5 epitope tag (to detect DUX4.V5) or eGFP. DUX4 protein was reduced in cells treated with CMV.H19 (35 ± 7%, P = 0.0007, N = 5 independent replicates), miR-675-5p mimic (36 ± 9%, P < 0.0001, N = 3), CMV.MIR675 (53 ± 9%; P < 0.0001, N = 4), U6.MIR675 (49 ± 8%; P < 0.0001, N = 5), and U6.MIR675-5p (42 ± 2%; P = 0.0004, N = 5). H1.MIR675 showed the highest DUX4 inhibition with an average of 91 ± 4% (P < 0.0001, N = 6), similar to U6.miDUX4.405 (93 ± 4%, P < 0.0001, N = 6). The U6.MIR675-3p construct did not significantly inhibit DUX4 protein levels (11 ± 7%, P = 0.6286, N = 5). b MiR-675 specifically reduced DUX4 expression via target engagement. MiR-675-resistant DUX4 constructs expressing full-length DUX4 protein or the Renilla luciferase-DUX4 reporter (CMV.DUX4-miR-675Res and RenLuc-DUX4.ORF-miR-675Res) contain silent mutations in miR-675 target site 780 (TS780) and lack miR-675 binding sites in the DUX4 3′UTR. Western blots show that miR-675 expression constructs did not significantly reduce miR-675-resistant DUX4 (P = 0.9997), while the U6.miDUX4.405 positive control, which had intact binding sites on DUX4, reduced mean DUX4 protein levels by 90 ± 6% (P = 0.0003). Similarly, increasing doses of the H1.MIR675 plasmid did not significantly silence the miR-675-resistant Renilla luciferase target (P = 0.3467 for the 14:1 miR-675:DUX4 ORF-miR-675Res ratio). Blots shown represent 6 (a) and 3 (b) independent blots (additional blots in Supplemental Data). DUX4 protein (DUX4p) was reported as mean % expression ± SEM and plotted above each representative image. Luciferase assay results (b) represent mean relative Renilla luciferase activity ± SEM, normalized to the miLacZ negative control (N = 3 independent experiments). For a, b one-way ANOVA followed by Dunnett’s multiple comparison tests were performed for statistical analyses. TS780WT: wild-type TS780 sequence. TS780M: TS780 sequence with silent point mutations colored in gray. ΔTS780: deleted TS780. Source data are provided as a Source data file.