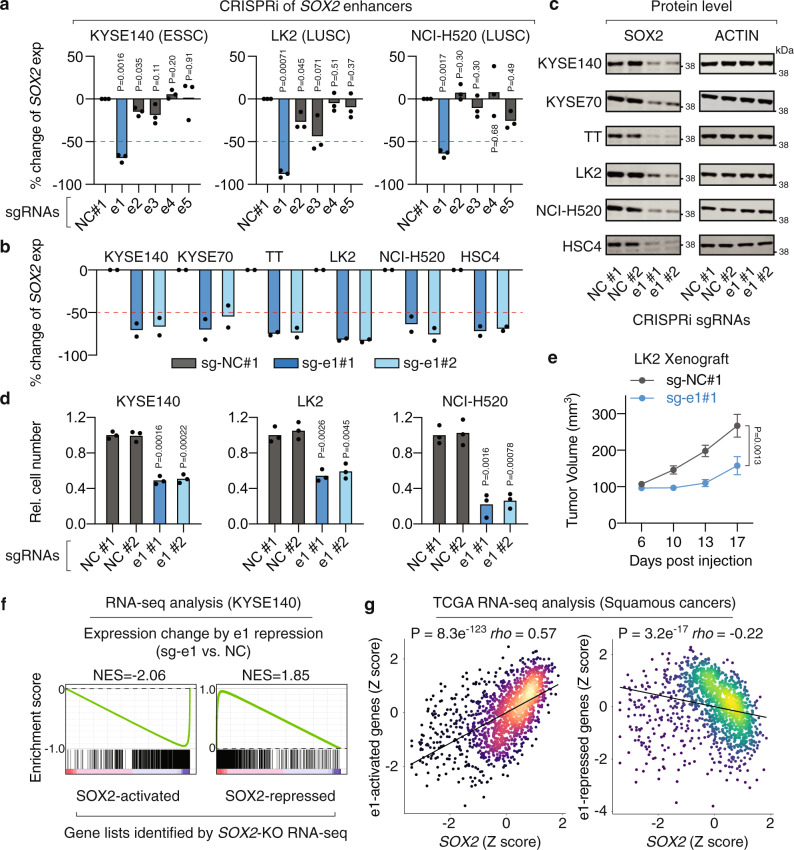
Fig. 3. Identification of a predominant single enhancer driving SOX2 overexpression.
a RT-qPCR measuring expression changes of SOX2 after CRISPR-mediated repression of each of the enhancers e1–e5. The expression level of SOX2 was normalized to cells treated with sg-NC#1 that has no recognition site in the genome. n = 3 biologically independent experiments. P values are derived from two-sided t tests. b RT-qPCR measuring expression changes of SOX2 after CRISPR-mediated repression of the e1 enhancer. Two separate sgRNAs sg-e1#1 and sg-e1#2 were used to target the e1 enhancer in six squamous cancer cell lines. n = 2 biologically independent experiments for each sgRNA. c Immunoblots showing protein level changes of SOX2 after repression of the e1 enhancer in six squamous cancer cell lines. The ACTIN protein serves as a loading control. The immunoblotting experiment was repeated once independently with similar results. d Cell proliferation of squamous cancer cell lines with and without e1 repression. Cell numbers were counted 6 days post seeding and normalized to the sg-NC#1 control. n = 3 biologically independent experiments. P values are derived from two-sided t tests. e In vivo tumor growth of xenografts that were derived from LK2 cells with and without e1 repression. Error bar: standard error of the mean. n = 10 biologically independent samples of each condition. P value was derived from a two-way ANOVA with repeated measures. f GSEA analysis measuring expression changes of SOX2-activated (left) and SOX2-repressed (right) genes, identified by RNA-seq assays in KYSE140 cells with and without SOX2 knockout, after e1 repression in KYSE140 cells (n = 2 biologically independent experiments). g Relationship between SOX2 and e1-activated (left) and e1-repressed (right) genes across TCGA squamous cancer samples. Z-scores were generated as described in the “Methods” section. P values were derived from two-sided Spearman’s correlation. Source data are provided as a Source Data file.