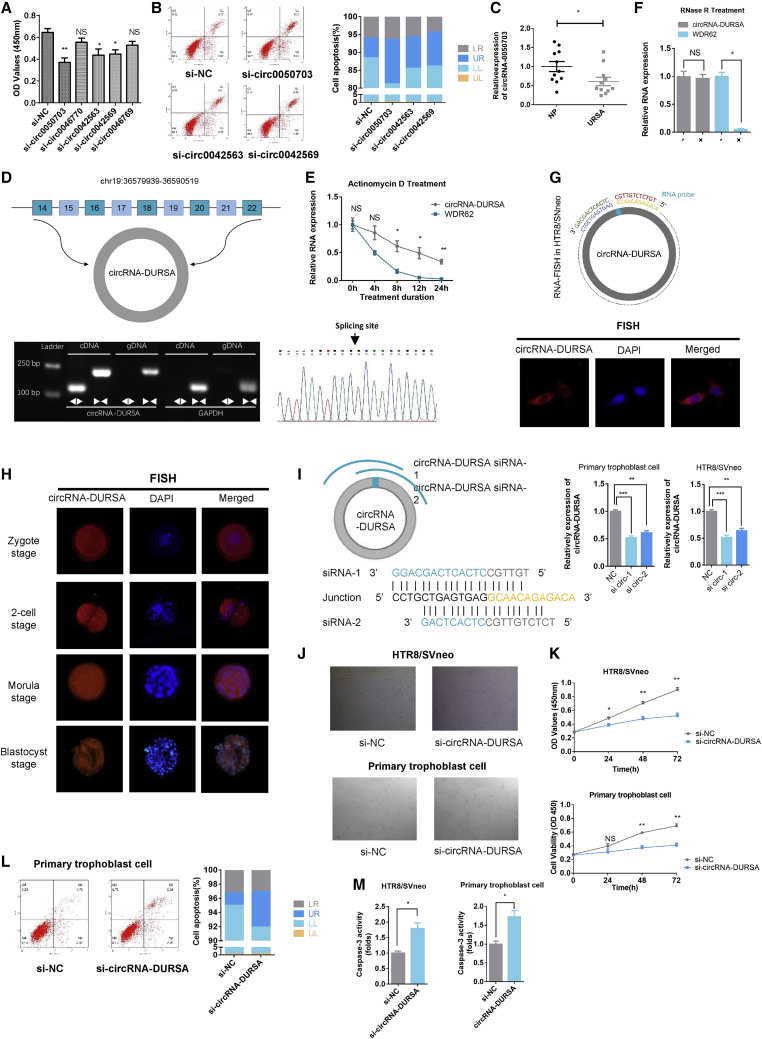
Figure 2.
Characterization of circRNA-DURSA
(A) CCK-8 assays of HTR8/SVneo cells transfected with circRNA-0050703, circRNA-0046770, circRNA-0042563, circRNA-0042569, or circRNA-0046769 siRNA. (B) Flow cytometric analysis of cell apoptosis in HTR8/SVneo cells transfected with circRNA-0050703, circRNA-0042563, and circRNA-0042569 siRNA. (C) Relative expression of circRNA-DURSA in villus tissues of normal pregnancy and URSA (n = 22). (D) (Top) Schematic diagram of genomic location and splicing of circRNA-DURSA. (Bottom left) RT-PCR products using divergent and convergent primers showing circularization of circRNA-DURSA. GAPDH was used as a negative control. (Bottom right) Arrow represents the “head-to-tail” splicing sites of circRNA-DURSA according to Sanger sequencing. (E) circRNA-DURSA resistance to actinomycin D (ActD) was detected by qRT-PCR. (F) circRNA-DURSA and linear WDR62 mRNA expression in HTR8/SVneo cells treated with RNase R. (G) RNA FISH showed the expression and localization of circRNA-DURSA in HTR8/SVneo cells. DAPI solution was used to stain the nuclei. (H) RNA FISH showed the localization of circRNA-DURSA in human pre-implantation embryos. (I) Two different siRNAs were designed for targeting the back-splicing junction of circRNA-DURSA. HTR8/SVneo cells and human primary trophoblast cells were transfected with si-circRNA-DURSA or si-circRNA-NC, and the expression of circRNA-DURSA was detected by qRT-PCR to select the most effective siRNA. (J) The experiments of (J)–(M) were carried out in the groups transfected with si-circRNA-DURSA or si-NC. Cell morphology was determined by microscopy. (K) CCK-8 assays were used to determine cell viability. OD values were obtained at 24, 48, and 72 h after transfection. (L) Flow cytometric analysis of cell apoptosis in treated primary trophoblast cells. (M) Caspase-3 activity was measured to determine the activation level of the caspase-3 protein. Three different independent experiments with three technical repetitions were performed. Data are expressed as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant.