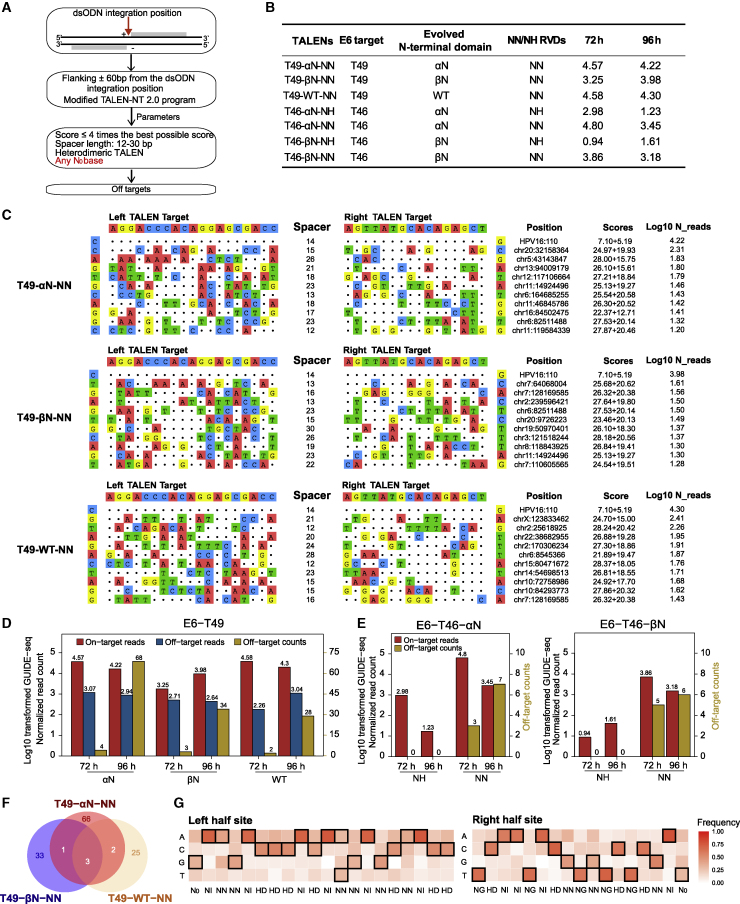
Figure 4.
TALEN off-target spectrum in HPV E6-targeted sites
(A) Flowcharts of TALEN off-target identification algorithm. (B) All TALEN variants targeted the HPV16 E6 gene. The engineered N-terminal domains, containing NN/NH RVDs and log10-transformed normalized GUIDE-seq on-target reads, are summarized for all variants. (C) Schematic of on-targets and top 10 off-targets of three T49 TALENs at 96 h. The on-target is at the top line followed by off-targets, with mismatches highlighted in color. The specific cleavage positions, TALEN-NT 2.0 scores of left and right TALEs, as well as log10-transformed normalized GUIDE-seq reads are displayed at the right of each site. (D) Bar plots of the log10-transformed normalized GUIDE-seq on-target reads, off-target reads, and off-target counts for each T49 TALEN. Both the 72- and 96-h data are shown. The double y axis represents log10-transformed normalized GUIDE-seq read counts (left) and off-target numbers (right). (E) Bar plots of the log10-transformed normalized GUIDE-seq on-target reads, off-target reads, and off-target counts for T46-αN-NN/NH and T46-βN-NN/NH TALENs. Both the 72- and 96-h data are shown. The double y axis represents log10-transformed normalized GUIDE-seq read counts (left) and off-target numbers (right). (F) Venn diagrams illustrating overlapped off-target of three T49 TALENs. Both the 72- and 96-h data were used. (G) Recognized nucleotide frequencies of each RVD derived from T49-WT-NN off-targets.