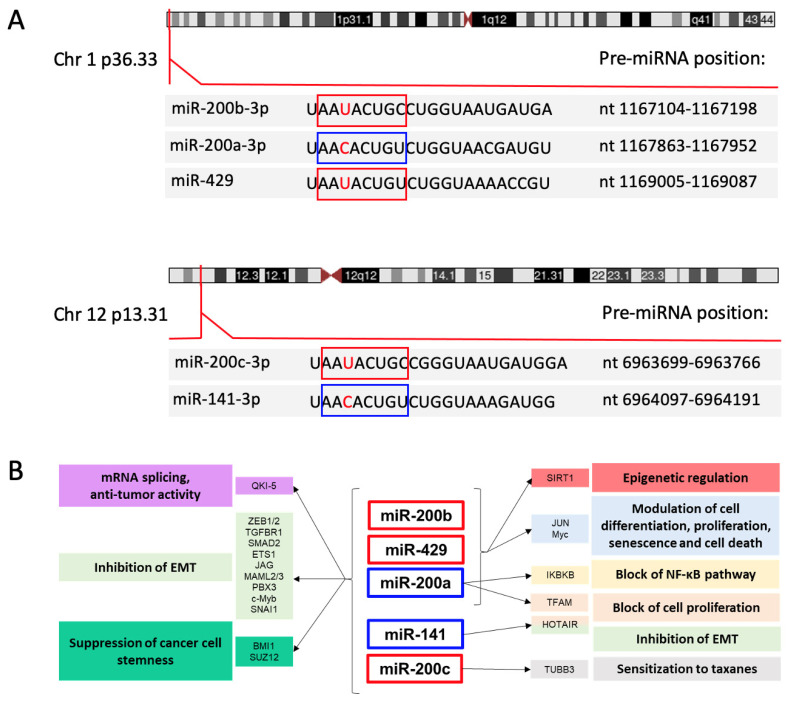
Figure 1.
Genetic organization of the miR-200 family in humans. (A) Indicated are the chromosome loci encoding the two clusters of the miR-200 family, the positions of the pre-miRNA hairpins, and the sequences of the mature miRNAs with seed sequences highlighted in boxes. The seed sequences differ by one nucleotide (indicated in red). As described in the text, there is a strong bias for production of the -3p products from the pre-miRNAs. Information is from the Sanger miRbase (https://www.mirbase.org, accessed on 28 July 2021) [12] and https://genome.ucs.edu/index.htlm (accessed on 11 October 2021) University of California Santa Cruz Genome browser. (B) Schematic representation of the main targets and pathways controlled by the miR-200 family members grouped according to their genomic clusters. Red and blue boxes indicate the miRNAs with the two different seed sequences. A selection of target genes and corresponding biological effects are indicated using matching colors.