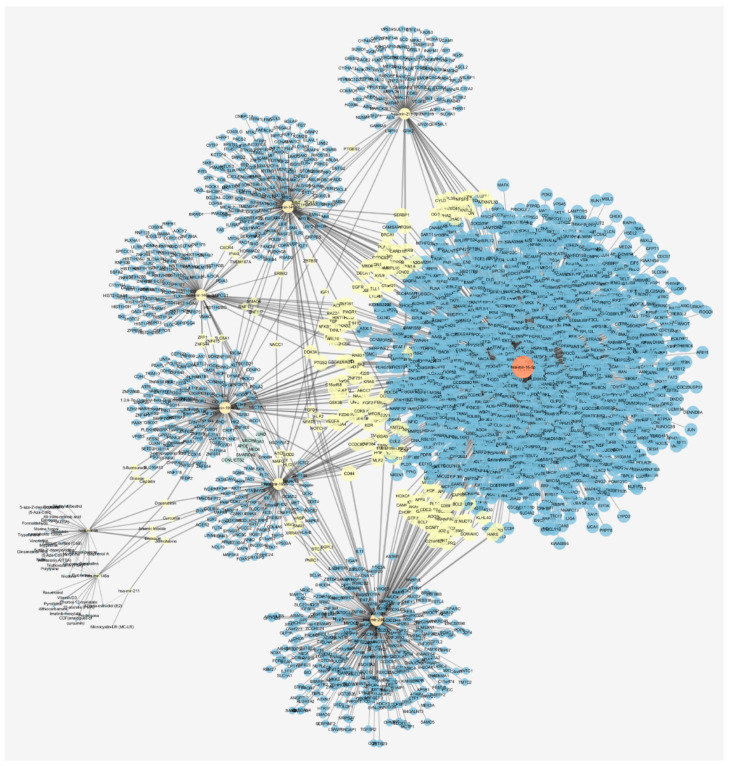
Figure 1.
Gene–miRNA interactions were collected from three well-annotated databases: miRTarBase (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php, 22 November 2021), TarBase v.8 (https://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex, 22 November 2021), and miRecords (http://c1.accurascience.com/miRecords/, 22 November 2021). The miRNA–compound interaction data were collected from SM2miR (http://www.jianglab.cn/SM2miR/, 22 November 2021) and PharmacomiR (http://www.pharmaco-mir.org/, 22 November 2021). Centrality network parameters (crucial for identification of nodes important for network stability) were calculated by using Cytoscape Network Analysis. Node dimension, which is proportional to closeness centrality values, and node color (blue < red) represents betweenness centrality values. Node thickness is proportional to edge betweenness values. Closeness centrality: distance of each node from all other nodes of the network; betweenness centrality: measure of the importance on a node basing on the shortest paths it is included into; edge betweenness: number of shortest paths between nodes that contain the edge.