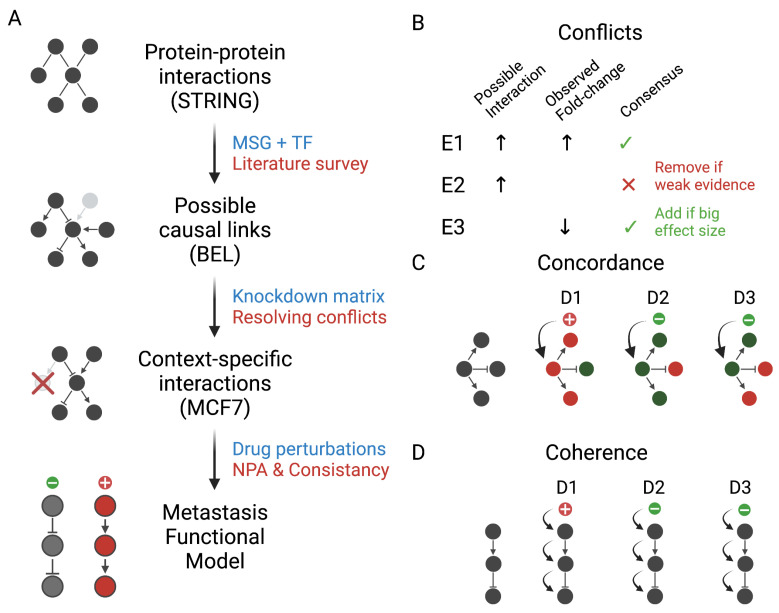
Figure 1.
A workflow for building, refining, and evaluating a breast cancer-specific network model of metastasis. (A) We queried the String database for protein–protein interactions for metastasis suppressor genes (MSG) and transcription factors (TF). Next, we surveyed the relevant literature to vet and supplement the interactions and express them as possible causal links in the biological expression language (BEL). Finally, we filtered and evaluated the network model using knockdown and drug perturbation datasets. (B) Conflicts between the data-driven and curated interactions were resolved by prioritizing the context, the strength of the evidence, and the effect sizes. We scored the metastasis model on drug perturbations using the network perturbation amplitude (NPA). We divided the network into smaller subgraphs and measured the agreement between the expected and the observed direction activity. We considered the consistency in (C) subnetworks of nodes connected to an upstream by one edge (concordance) and (D) paths connecting a particular node to its upstream by a sequence of edges (coherence).