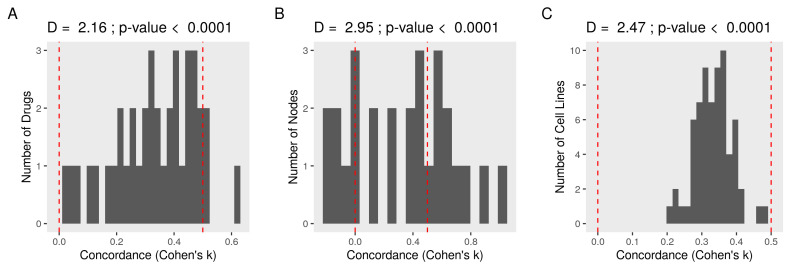
Figure 5.
Concordance of expectations and observations in the subnetworks of the metastasis model. We calculated and binarized (1 for activation or −1 for repression) the perturbation coefficients of every node in the network. We then evaluated the agreement between the expected and observed direction of change in the subnetworks of nodes connected to an upstream by one edge (concordance). First, we multiplied the drug’s effect on the upstream node by the sign of the edges to form expectations. Next, we compared the expectations with the actual perturbation coefficients of the corresponding nodes. Negative Cohen’s indicates worse and positive better agreement than expected by chance. Finally, we compared the probability distribution of the concordance to randomly generated values using Kolmogorov-Smirnov (KS) test. D is the maximum distance between the cumulative distribution functions (ECDF). (A) A histogram of the average concordances for every drug. (B) A histogram of the average concordances for every node. (C) A similar workflow was applied to other cancer cell lines (n = 67), and the concordance values averaged by cell lines are shown as a histogram.