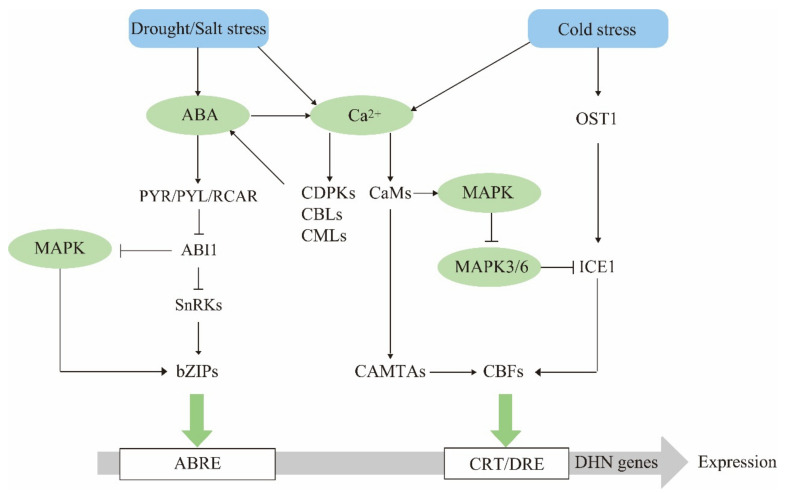
Figure 1.
Dehydrin gene expression regulatory network model. Abiotic stressors such as drought, high salinity, and cold trigger dehydrin expression in plants depending on various signaling pathways. Drought and high salinity activate the ABA and calcium signaling pathways. After ABA binds its corresponding receptor, it acts on bZIP TFs and mediates dehydrin expression. Calcium ions are decoded by proteins such as CaMs, CDPKs, CBLs, and CMLs and regulate dehydrin expression via CBF TFs. They also act as bridges connecting drought/salinity and cold stress responses. ICE1 protein occurs in the early cold stress pathway. The CBFs in this pathway regulate the expression of multiple downstream CORs. The MAPK cascade also occurs throughout the entire regulatory network and regulates dehydrin expression upstream. Arrows and bars indicate positive and negative regulation, respectively.