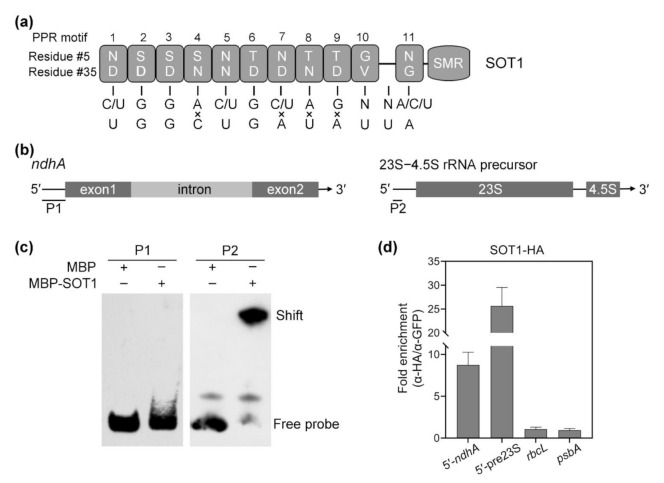
Figure 4.
SOT1 binds the 5′ region of the ndhA transcripts in vivo. (a) Schematic illustration of the PPR motif in SOT1 and its alignment with the predicted binding site in the 5′ region of the ndhA transcripts. Each box indicates the amino acid residues at positions 5 and 35 of an individual PPR motif. The potential nucleotide targets of PPR motifs are shown below. (b) Schematic representation of the domain structures of ndhA mRNA and the 23S−4.5S rRNA precursor. The positions of probes P1 and P2, which were subjected to electrophoretic mobility shift assays (EMSAs), are shown below the models. (c) EMSA showing that SOT1 alone exhibits little binding activity to the 5′ ends of ndhA transcripts in vitro. A total of 150 nM recombinant MBP and MBP-SOT1 proteins were incubated with 10 nM biotin-labeled probes (P1 and P2). Three independent experiments were performed, and one representative experiment is shown. (d) RNA coimmunoprecipitation assays identify that SOT1 binds the 5′ region of ndhA transcripts in vivo. The intact chloroplasts were isolated from 12-day-old sot1-3 complemented plants (sot1-3/35S:SOT1-HA). The chloroplast extracts were subjected to immunoprecipitation against HA and GFP antibodies. SOT1 was reported to bind the 5′ ends of the 23S−4.5S rRNA precursor [22,23]; therefore, this interaction served as a positive control. The sample immunoprecipitated with the GFP antibody served as a negative control. The relative RNA-enrichment levels were determined using qPCR. 5′-ndhA and 5′-pre23S indicate the 5′ ends of ndhA transcripts and 23S−4.5S rRNA precursor, respectively. Mean values ± SD of the triplicate replicates are shown.