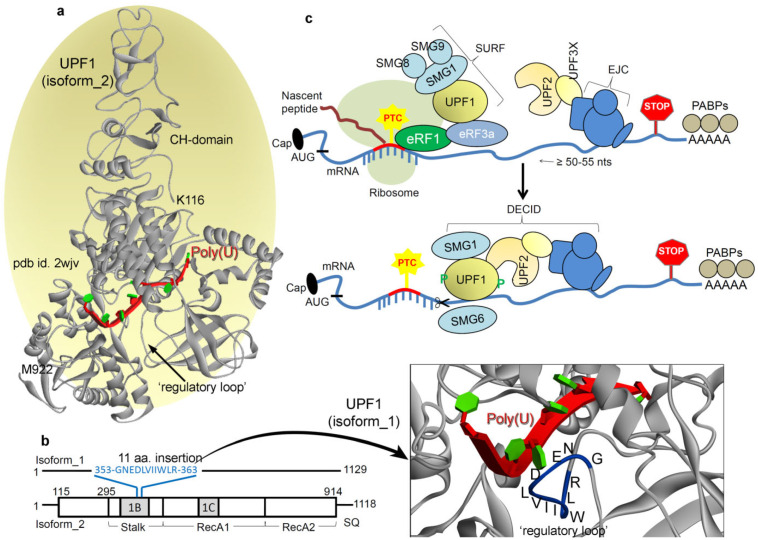
Figure 1.
The master regulator of the NMD (nonsense-mediated mRNA decay) pathway: UPF1 (UP-frameshift 1). (a) Optimized UPF1 isoform_2 model structure [40] with the poly(U) mRNA motif, constructed based on the pdb (protein data bank) id: 2wjv [67] and 2xzo [37]. The crystal structure of UPF1 (pdb id: 2wjv [67]) is with the UPF2 protein, but since UPF1 adopts a different conformation when bound with UPF2, we removed it from the presentation. In addition, the poly(U) mRNA motif has been inserted into the structural representation (from pdb id: 2xzo [37]) as a reference since it is not present in the original crystal structure (pdb id: 2wjv [67]). (b) Isoform_1 structure of the UPF1 (build considering the pdb id: 2wjv [67]) protein, with the 11 aa (amino acids) insertion (353-GNEDLVIIWLR-363) in the ‘regulatory loop’. These 353-GNEDLVIIWLR-363 residues are highlighted over the protein structure in blue color. This UPF1 isoform_1 structure with inserting 11 aa was modeled considering the isoform_2 crystal structure as the template (pdb id: 2wjv) [67]. (c) In the NMD pathway, recognition of PTC-containing mRNA transcript requires UPF1, UPF2, and UPF3 proteins as the core machinery, along with various suppressors with morphogenetic effect on genitalia (SMG) proteins [7,61]. There is a general agreement that NMD substrate recognition relies upon differences in mRNA ribonucleoprotein (mRNP) composition between normal mRNAs and PTC-containing mRNA transcripts. In mammalian cells, an exon–exon junction complex (EJC) of proteins deposited at exon–exon boundaries during pre-mRNA splicing is considered to be a primary determinant of NMD [62]. Translation termination at a nonsense codon located ~50–55 nucleotides upstream of an exon–exon junction usually triggers NMD. It is suggested that translation termination events leading to NMD entail the SURF complex, consisting of SMG1, UPF1, eRF1 (eukaryotic translation termination factor 1), and eRF3 [41]. Once translation terminates, UPF1 then interacts with UPF2 that is bound to the EJC together with additional UPF3 proteins. These interactions result in the assembly of the surveillance complex, UPF1 phosphorylation by SMG1, and eventually mRNA degradation [63]. In principle, if an EJC-related exon–exon junction resides ≥50–55 nucleotides downstream of the termination event, then NMD initiates. Concerning the dynamics of NMD components, the binding of UPF2 with UPF1 causes a large conformational change in the UPF1 CH-domain (cysteine-histidine-rich domain), which triggers the UPF1 helicase activity. Furthermore, within the resulting decay-inducing complex (DECID), the SMG1 protein phosphorylates UPF1 that inhibits further translation termination initiation at the AUG codon [56].