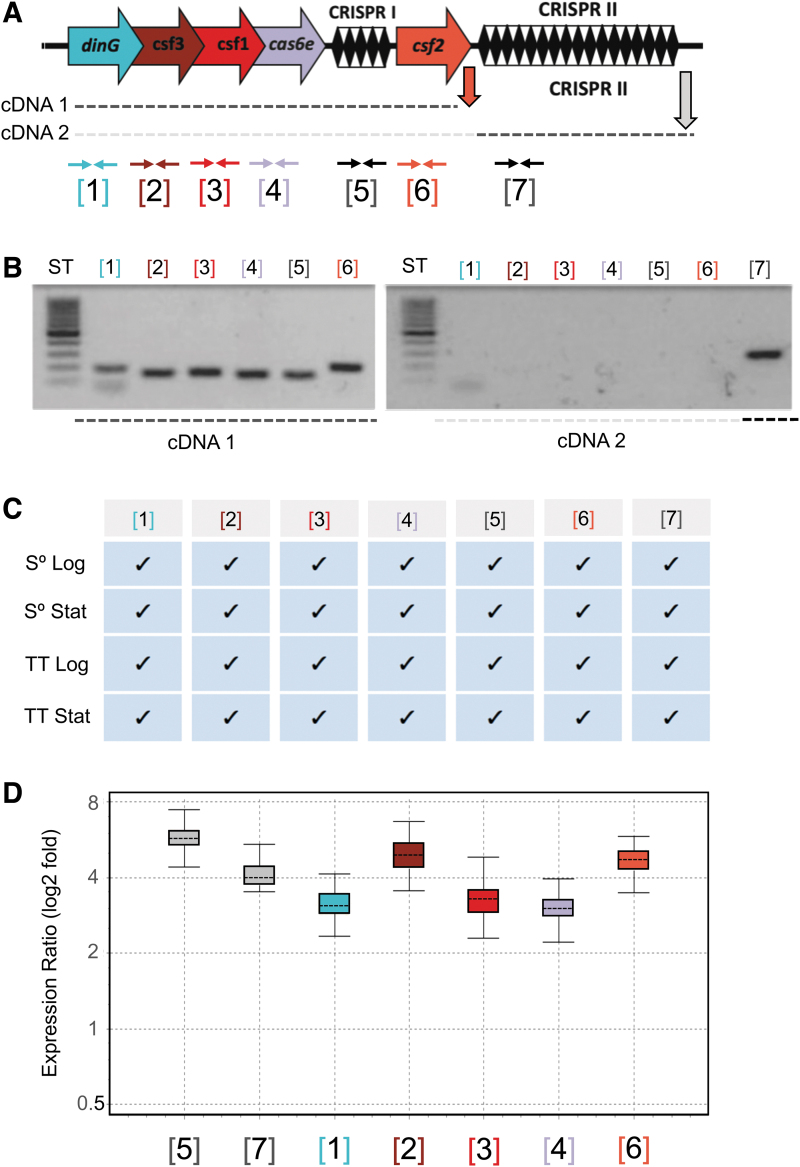
FIG. 4.
(A–D) Expression of the type IV CRISPR-Cas system of F. caldus ATCC 51756T. Generic scheme showing the F. caldus CRISPR-Cas system, the cDNAs obtained by reverse transcription (dotted lines), and reverse transcription (RT) polymerase chain reaction (PCR; vertical arrows) and PCR primers position (horizontal arrows), numbered according to the position of the different features in the loci. Primers used in each reaction are detailed in Supplementary Table S2. (A) Analysis of the RT-PCR-amplified products by agarose gel electrophoresis. Total RNA was extracted from mid-log sulfur-grown cultures (pH 2.5; 40°C, 150 rpm; mineral salts medium) and used as template in cDNA synthesis with reverse primers indicated in panel (A). (B) Summary of RT-PCR amplification results for total RNA extracted from cells grown under different growth conditions. Cultures were grown in the presence of either sulfur (S°) or tetrathionate (TT) as energy sources, and cells were collected at two distinct growth phases, logarithmic (Log) and stationary (Stat). Positive amplification is indicated by a checkmark. (C) Fold induction of each component as assessed by quantitative RT-PCR for total RNA recovered from sessile (biofilm) versus planktonic grown cells (achieved in 6 cm long and 2.5 cm diameter columns containing elemental sulfur-quartz). Gene expression levels in each condition were normalized against a reference gene (rpoC, “housekeeping”), and normalized values were used in the calculation of the expression ratio sessile/planktonic.