Figure 7.
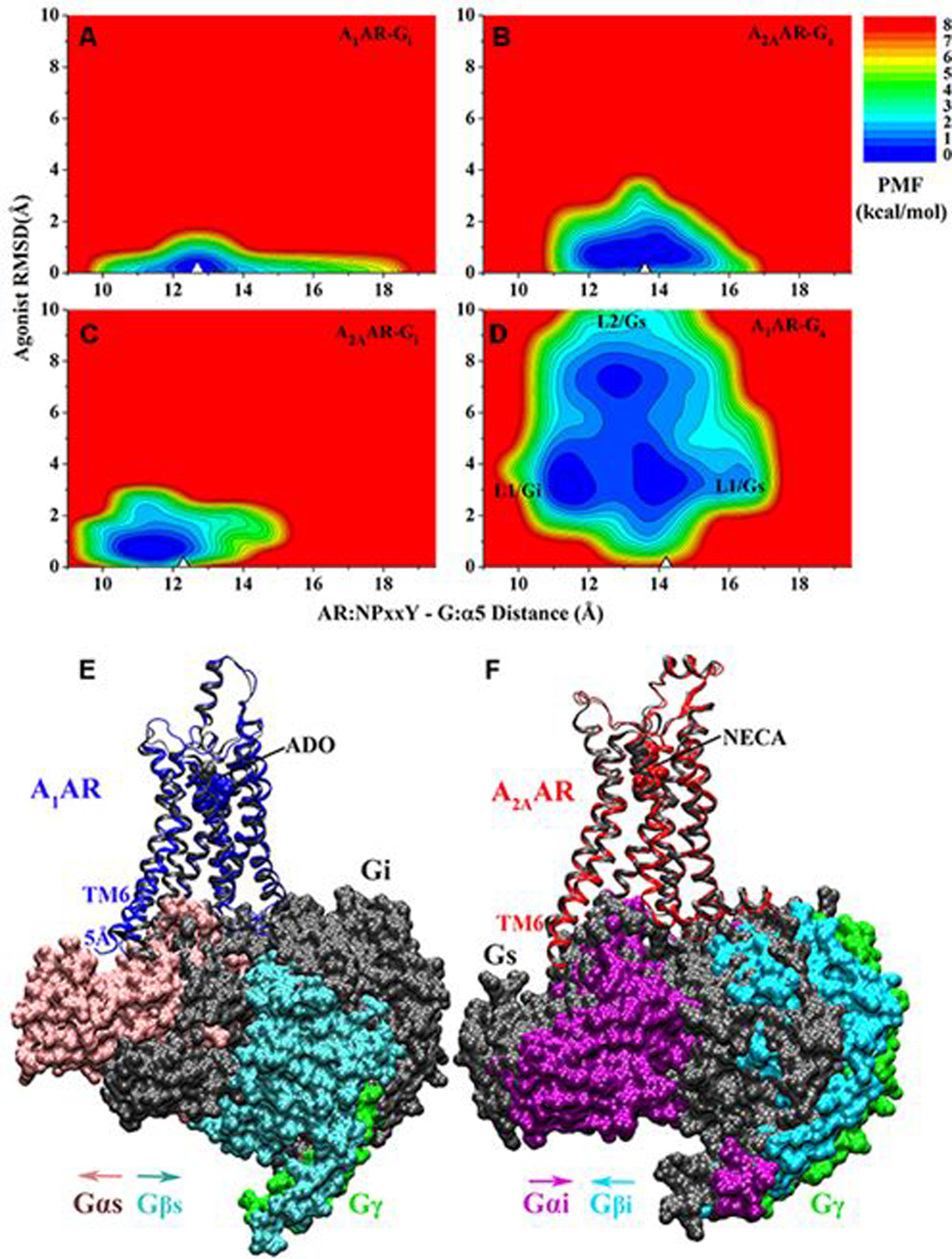
2D potential of mean force (PMF) profiles of the (A) ADO-bound A1AR-Gi, (B) NECA bound A2AAR-Gs, (C) NECA bound A2AAR-Gi and (D) ADO bound A1AR-Gs complex systems regarding the agonist RMSD relative to the cryo-EM conformation and AR:NPxxY-G:α5 distance. The white triangles indicate the cryo-EM or simulation starting structures. Summary of specific AR-G protein interactions: (E) the ADO-bound A1AR prefers to bind the Gi protein to the Gs. The latter could not stabilize binding of agonist ADO in the A1AR and tended to dissociate from the receptor. (F) The A2AAR could bind both the Gs and Gi proteins, which adopted distinct conformations in the complexes. Adapted with permission from Wang et al. (2019). Copyright 2019 American Chemical Society. https://pubs.acs.org/doi/10.1021/acs.jpcb.9b04867. Further permissions related to the material excerpted should be directed to the American Chemical Society.
