Fig. 1 |. Different mechanisms of autophagy.
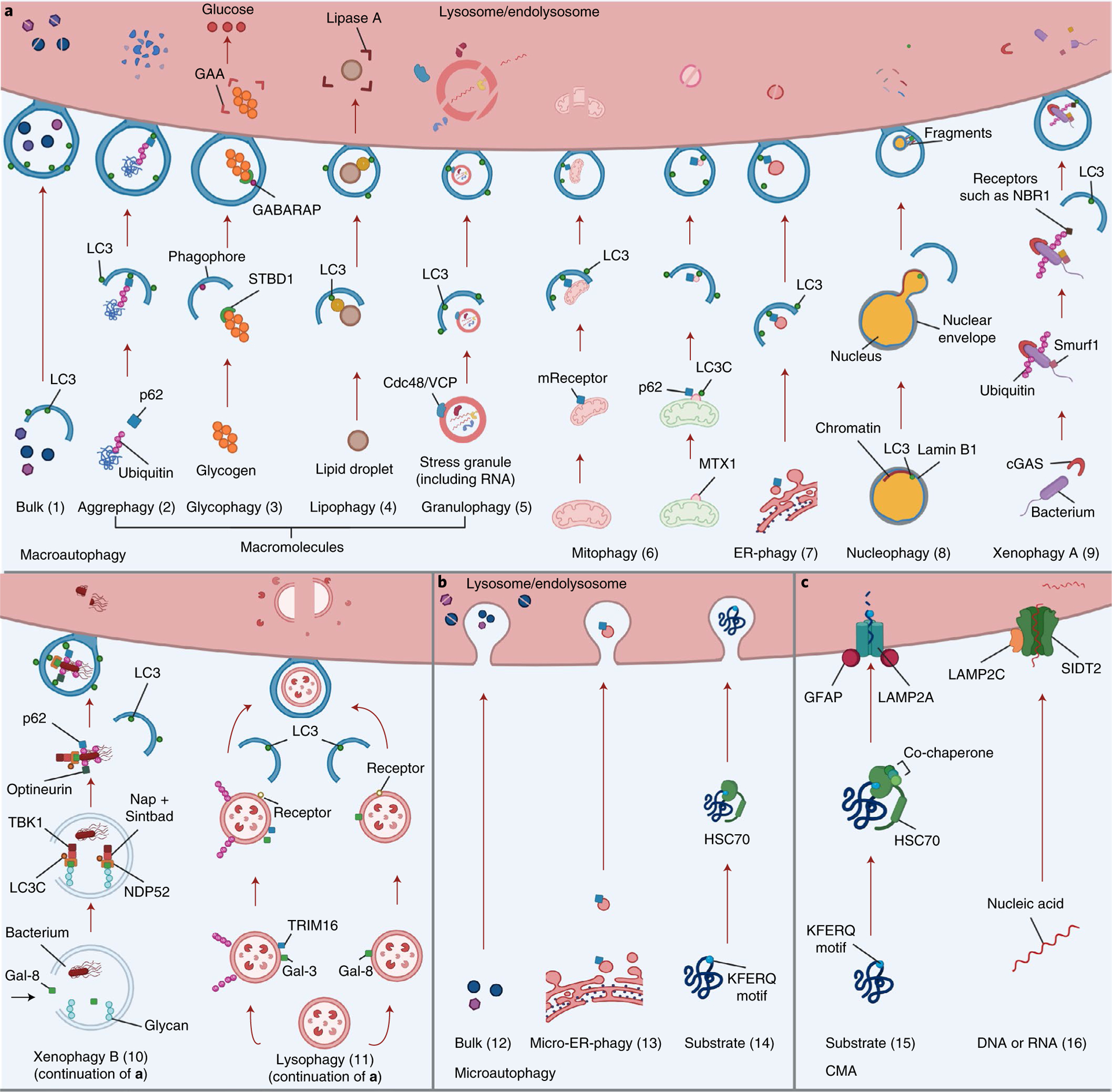
a, Macroautophagy (referred to herein as autophagy) (1) is a nonselective process that targets macromolecules or subcellular organelles in bulk. Cytoplasmic material is sequestered into an autophagosome and delivered to the lysosome (or endolysosome) for degradation). Selective autophagy involves recognition of specific cytoplasmic cargo via autophagy receptors that also interact with LC3 in the autophagic membrane, leading to cargo sequestration into autophagosomes that are delivered to a lysosome (or endolysosome) for degradation. This includes aggrephagy (2), where aggregated proteins are ubiquitinated and targeted by ubiquitin-binding autophagy receptors such as p62 (or NBR1); glycophagy (3), where STBD1 (genethonin-1) binds to glycogen and GABARAP, facilitating lysosomal glycogen breakdown into non-phosphorylated glucose by enzymes such as GAA; lipophagy (4), in which lysosomal lipids are degraded into free fatty acids, which are then converted into ATP; the identity of the receptor(s) (yellow) involved in sequestration of lipid droplets is unknown;; granulophagy (5), where sequestration of stress granules (RNA + protein) is mediated by Cdc48/VCP, allowing the stress granule to be delivered to the lysosome for degradation; mitophagy (6), where damaged mitochondria are bound by soluble or membrane-bound mitophagy receptors (mReceptors) that can also bind LC3, leading to engulfment of the mitochondrion into an autophagosome and subsequent delivery to a lysosome for degradation (left); in piecemeal mitophagy, degradation of parts of mitochondria occurs via binding of the outer mitochondrial membrane protein metaxin-1 (MTX1, in the extruded fraction) to LC3C, resulting in the recruitment of p62 and autophagosome formation (right); ER-phagy (7), which in mammals uses the specific receptors FAM134B, RTN3L, ATL3, SEC62, CCPG1 and TEX264, which are located in different parts of the ER; these receptors bind to LC3, leading to sequestration of the ER into an autophagosome and lysosomal degradation of the ER; nucleophagy (8), which, when triggered in mammals, results in nuclear LC3 binding to lamin B1, leading to formation of a bulge that is pinched off to the cytoplasm where degradation by autophagy occurs; xenophagy (type A, 9), where a bacterium’s DNA is detected by cGAS, a sensor that triggers a process of ubiquitination via Smurf1; this is followed by attachment of the NBR1 receptor to the ubiquitin chains and LC3 to continue the autophagy process for degradation of the bacterium; xenophagy (type B, 10), where a bacterium damages the membrane of the phagosome, exposing interior glycans that recruit galectin-8 (Gal-8), which is then recognized by NDP52 to recruit TBK1, LC3C, Nap and Sintbad; the optineurin, p62 and NDP52 receptors interact with ubiquitin on the pathogen and recruit the autophagic engulfment system, and the engulfed pathogen is then brought for degradation; and lysophagy (11), which occurs upon lysosomal membrane permeabilization and can be achieved with or without ubiquitination: recruitment of galectin-3 (Gal-3) to damaged lysosomes further recruits TRIM16 and autophagic proteins such as ULK1 and ATG16L1, and ubiquitination on the lysosome results in the recruitment of p62, which binds to LC3 to facilitate the autophagic process (left); in a parallel ubiquitin-independent process, galectin-8 is recruited to damaged lysosomes and is capable of directly binding to the NDP52 receptor that interacts with LC3 to continue the autophagic process (right). b, Microautophagy involves capture of cytoplasmic components through direct invagination of endolysosome membranes and can be nonspecific (bulk) (12) or highly specific (13,14). Examples of selective microautophagy in mammalian cells include micro-ER-phagy (13), which uses the SEC62 receptor and involves ER capture and degradation by invagination of the lysosome/endolysosome, and endosomal microautophagy of proteins with the KFERQ pentapeptide motif (14) in a process requiring the chaperone HSC70. c, CMA (15) also involves targeting of proteins containing a KFERQ pentapeptide-related motif by HSC70 and other co-chaperones such as HSP40. The substrate is then imported into the lysosome through the LAMP2A receptor for further degradation. The LAMP2A receptor is modulated by the glial fibrillary acidic protein (GFAP). Finally, in a CMA-like manner, DNAutophagy/RNAutophagy (16) can occur: nucleic acids (DNA or RNA) bind to the LAMP2C receptor (orange), which also binds to lysosomes. This process allows nucleic acids to be taken up by the lysosome. It has been proposed that a transporter called SIDT2 (green) might have a role in direct uptake of nucleic acids by the lysosome.
