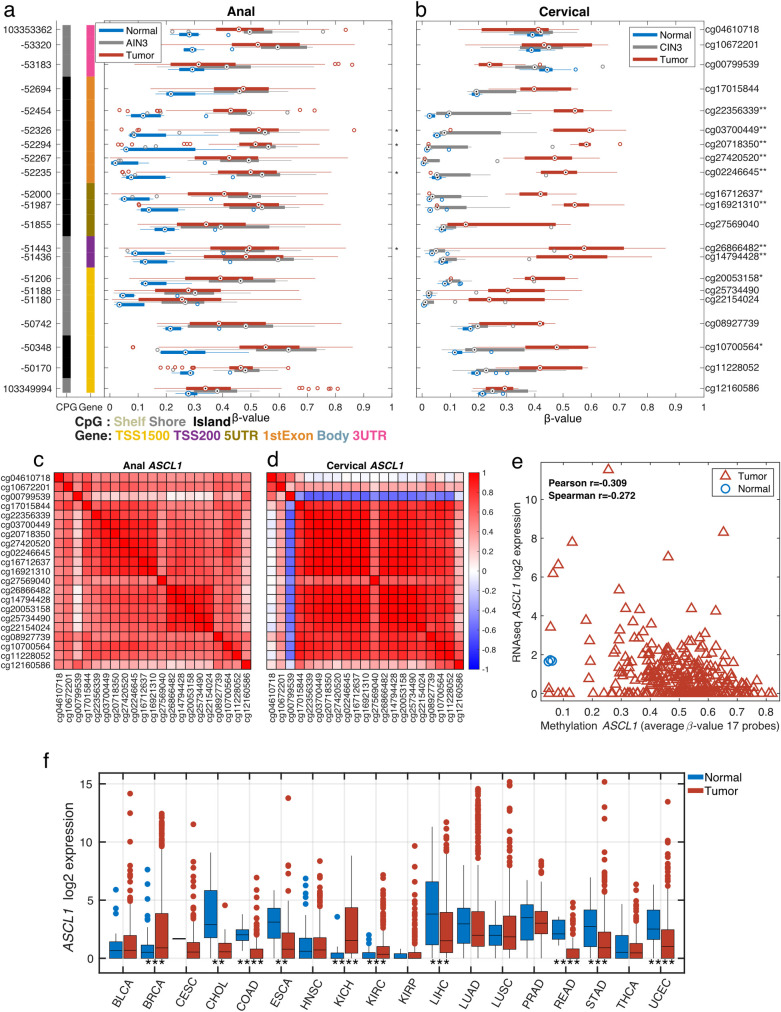
Fig 3.
Gene structure methylation plot of differentially methylated CpG loci within ASCL1 in anal (a) and cervical (b) tissues. Genomic coordinates are represented on the left vertical axis and methylation probe ID (or CpG loci) on the right vertical axis. For each of the 21 CpG loci, boxplots illustrate the median (dot) and interquartile ranges [25th (low boundary of box) and 75th (upper boundary of box) percentiles] of β-values in tumor (red boxes), AIN3 or CIN3 (blue boxes) and normal (green boxes). Significantly different median methylation at each CpG loci is noted as * for p<0.05 and ** for p<0.005. Among the 21 CpG sites presented, 4 CpG sites fall within the overlapping DMRs that were significantly hypermethylated in both anal (a) and cervical (b) cancers. For ASCL1, anal in situ (AIN3) showed similar methylation levels as tumors while, CIN3 methylation levels were similar to normal cervical tissues. Corresponding correlation plots between all ASCL1 probes for anal (c) and cervical (d) cancers show a high degree of correlation for all of the probes. The average methylation levels for 17 probes show low correlation (r = -0.3) to RNAseq gene expression level in the TCGA CESC dataset (e). The tumor vs. normal gene expression levels across multiple TCGA tumor types are shown (f, *p<0.05, **p<0.01, ***p<0.001 & ****p<0.0001).