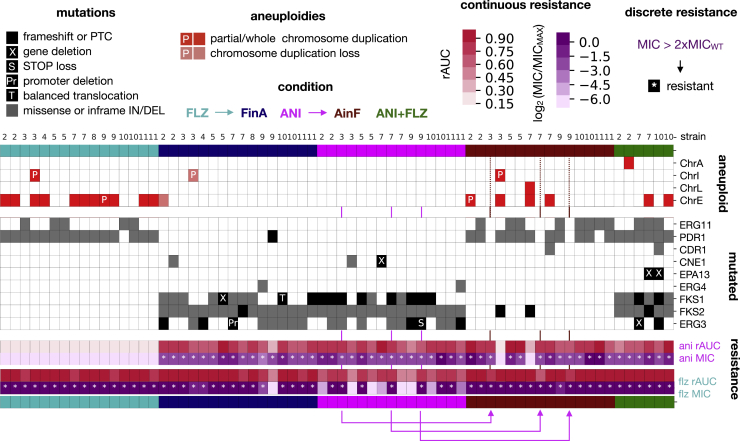
Figure 6.
Aneuploidies and recurrently mutated genes
Each drug is associated with a particular set of mutated genes and aneuploidies. Columns represent the evolved samples, each strain indicated by a number: 2, CST34; 3, EB091; 4, CST78; 5, M12; 6, EF1237; 7, EF1620; 8, F15; 9, CBS138; 10, P35; 11, BG2. Replicates of the same strain appear in the same order as in the experimental plate. Colors indicate the experimental condition. Blocks show, from top to bottom, chromosomal alterations, mutated genes, and susceptibility data. Whole and partial (P) chromosomal duplications appearing newly in each condition are marked as red, while losses are marked as light salmon boxes. Protein-altering mutations (gray boxes) and losses (black boxes) of genes appearing in at least 2 drug-evolved samples are shown. Note that we found a balanced translocation in FKS1 (T) and a deletion in the ERG3 promoter region (Pr) (Figure S3; Results; STAR Methods). PTC stands for premature termination codon. Pink arrows indicate the parent-daughter relationships for 3 AinF samples that did not present any new alteration in recurrent genes. Note that Figures S3 and S4 and Data S3 provide additional information about these mutations and genomic rearrangements. In addition, Figure S6 shows the association between these mutations and fitness or drug-resistance levels.