Figure 7.
ERG3 mutations and multidrug resistance
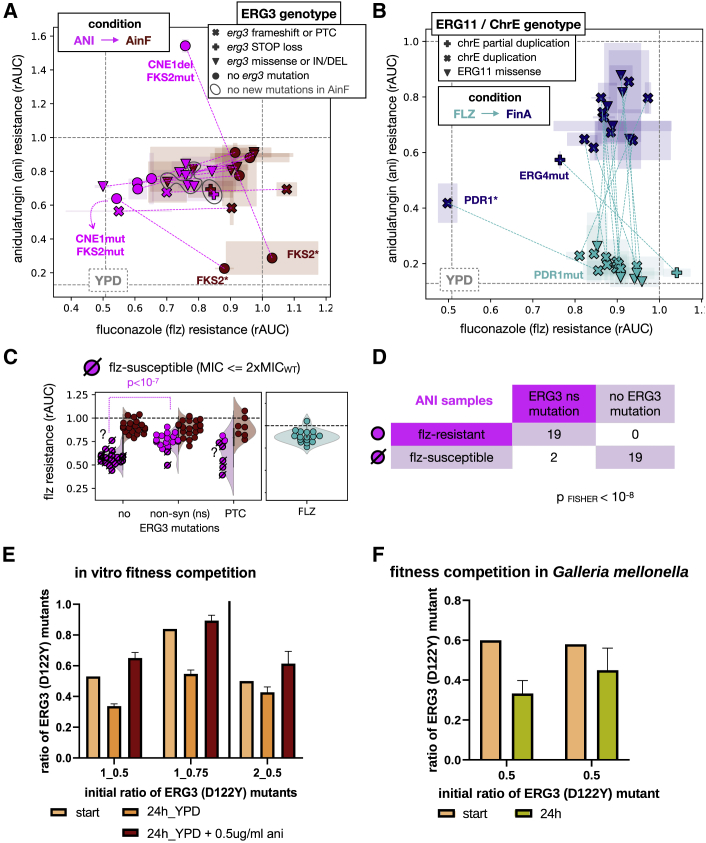
(A) Biplot showing the relationship between resistance (rAUC) toward ani and flz for a series of ANI/AinF related samples. The gray dashed lines indicate the rAUC = 1.0 (where fitness is maintained across the range of concentrations; Figure 1A) and the median rAUC across YPD samples for each of the 2 drugs. Each sample is represented by a symbol, with the color indicating the sample type: ANI (pink) and AinF (red) samples. The pink dashed lines indicate parent-daughter relationships (ANI-AinF) between the samples. The symbols represent different types of ERG3 mutations, and the gray circles outline 3 samples that did not acquire any new mutation in the recurrent genes in AinF. The 2 ANI samples with alterations in CNE1, which lost ani resistance due to truncations in FKS2(∗) in AinF samples, are marked. One of the ANI samples showed high ani resistance (above 1.0, meaning the fitness was higher in ani than in no drug), but also showed low basal fitness, which means that the high resistance value may be not representative. Error bars reflect the median absolute deviation across technical replicates.
(B) Relationship between rAUC of ani and flz in FLZ (light blue) and FinA (dark blue) samples. The green dashed lines indicate parent-daughter relationship (FLZ-FinA). The gray dashed lines indicate the rAUC = 1.0 (where fitness is maintained across all the range of concentrations; Figure 1A) and the median rAUC across YPD samples for each of the 2 drugs. No acquisition of ani resistance was observed in FLZ samples but only as a result of ani (FinA). The symbols represent the presence of ERG11 missense mutations or chromosome E aneuploidies. Two FinA samples showed a drop in flz resistance levels. One of them carried a PDR1 premature termination codon (∗), which resulted in susceptibility according to our MIC-based thresholding (STAR Methods) and reduced flz resistance below the median rAUC value of YPD samples. The other sample carried ERG4 mutation that resulted in a reduction but not a total loss of flz resistance. Error bars reflect the median absolute deviation across technical replicates.
(C) Non-synonymous (including missense and STOP loss) ERG3 mutations are associated with higher flz resistance (rAUC) in ANI samples. The p value corresponds to a Kolmogorov-Smirnov test. The corresponding AinF and FLZ samples are also shown for comparison of flz-resistance levels. The dashed symbols represent samples that were found to be flz susceptible according to our MIC-based thresholding (STAR Methods). Note that 2 samples (marked with “?”) were found as susceptible but have rAUC values in the range of resistant samples. This mismatch is clarified in Figures S7C and S7D. In addition, see Tables S2 and S3 for further information on the ERG3 mutations found in each sample.
(D) The presence of ERG3 non-synonymous mutations is correlated with discrete flz resistance in ANI samples. The number of ANI samples in each category and the p value of a Fisher test are shown.
(E and F) Growth competition between ani-resistant strains with and without ERG3 mutation (note that Table S5 includes the oligos used for sequencing). The y axis presents the calculated ratio of a sample with mutated ERG3 gene and the x axis ratios aimed at the beginning of the experiments. The error bars represent the standard deviation across technical replicates. (E) In vitro fitness competition of 2 pairs of strains: 1-CRISPR transformant ERG3 (D122Y) versus CRISPR transformant ERG3(WT) with NAT1 and 2-CRISPR transformant ERG3 (D122Y) with NAT1 versus 3H_ANI (ERG3 WT). The competition was conducted over a 24-h period and in YPD and YPD supplemented with 0.5 μg/mL ani in triplicates. (F) Two independent competition experiments in vivo. The fungal burden was obtained from 3 separate larvae for each of the initial mix of populations.