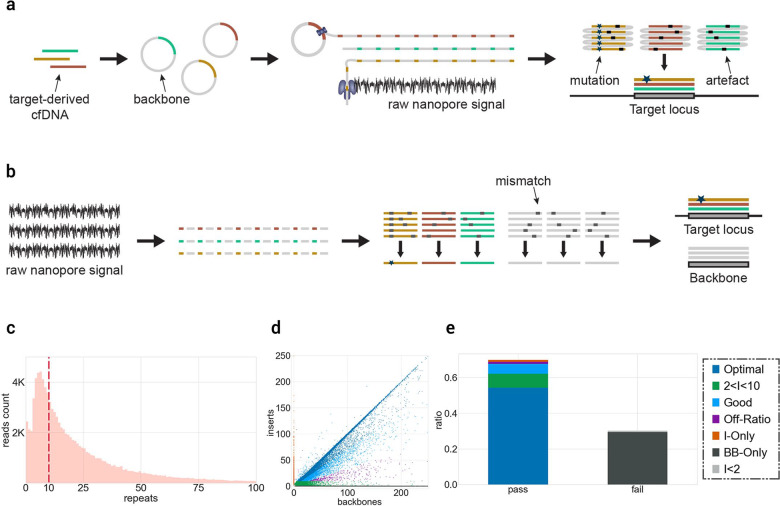
Fig. 1. CyclomicsSeq protocol.
a Experimental setup of CyclomicsSeq. PCR-amplified (‘target-derived') cfDNA is circularized with an optimized DNA backbone. Rolling circle amplification generates a long DNA molecule with alternating insert and backbone sequences, which is sequenced using ONT sequencing. Consensus calling of the DNA sequence allows discrimination between mutations and sequencing artifacts. b Schematic overview of the bioinformatic pipeline. c Distribution of insert copies versus the number of reads for a representative CyclomicsSeq run (#CY_SM_PC_HN_0002_001_000). d Ratio of insert versus backbone for CyclomicsSeq reads for a representative CyclomicsSeq run (#CY_SM_PC_HN_0002_001_000). Each read is represented by a data point (dot). Colors, noted in the legend, represent the different categories a read can belong to. Optimal CyclomicsSeq reads result from a one to one ratio of insert and backbone copies and contain at least 10 repeats (Blue). The other categories include: reads with fewer repeats (Green), reads without a backbone (Orange), reads without the target insert (Gray). Reads with BB:I ratios between 0.35 and 3 are defined as “Good” (Cyan), while the others are classified as “Off-ratio” (Purple). e Ratio of sequencing data grouped by read type for a representative CyclomicsSeq run (#CY_SM_PC_HN_0002_001_000). In this case, more than 60% of the data was used to generate consensus reads. The remnant data was discarded because it contained backbone-only sequences.