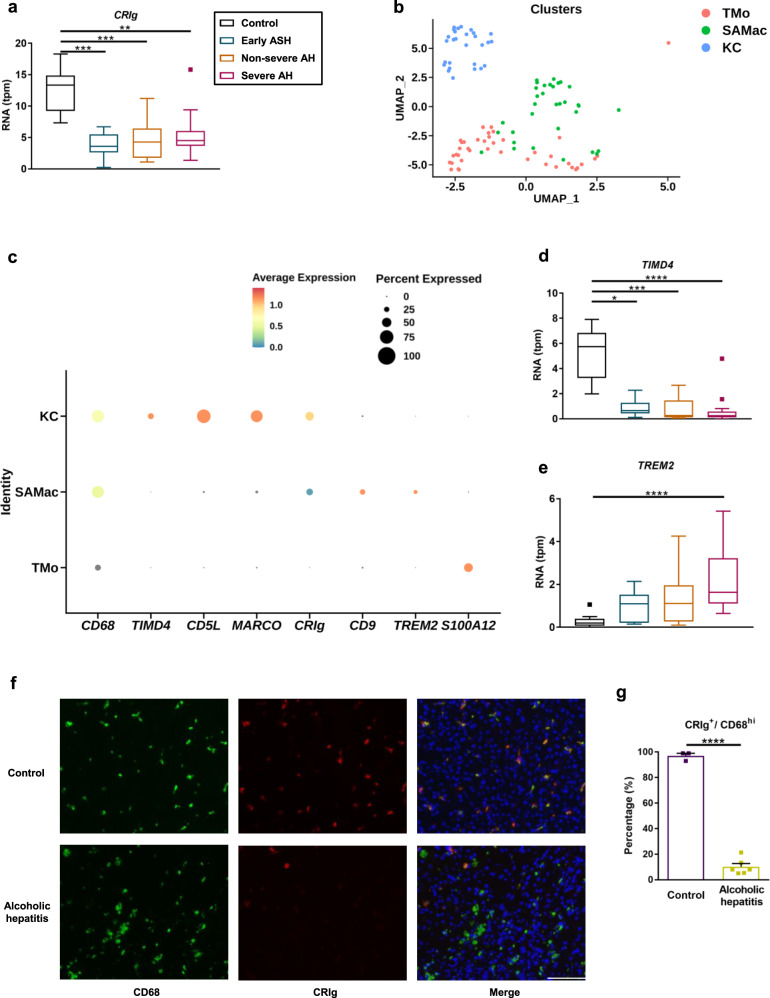
Fig. 1. Patients with alcohol-related liver disease have less CRIg expression on liver macrophages.
a, d, e RNA sequencing data of liver tissues from subjects without liver disease (Control; n = 10), patients with early alcoholic steatohepatitis (Early ASH; n = 12), non-severe alcoholic hepatitis (Non-severe AH; n = 11) and severe alcoholic hepatitis (Severe AH; n = 18). b and c Single-cell RNA sequencing data of liver tissues from subjects without liver disease (uninjured; n = 5) and alcohol-related cirrhotic patients (cirrhotic; n = 2). a mRNA level of CRIg. b UMAP annotating different macrophage cell types as KC (Kupffer cell), TMo (tissue monocyte) and SAMac (scar-associated macrophage). c Dot plot showing expression of CRIg and other stated marker genes in human liver macrophage subpopulations. d mRNA level of TIMD4 (marker for Kupffer cells). e mRNA level of TREM2 (marker for scar-associated macrophages). f Representative liver sections of CD68 and CRIg immunofluorescence staining. g Quantification of the stained liver sections. Scale bar = 100 μm. Results are expressed as median with interquartile range (a, d, e), or mean ± s.e.m. (g). P value is determined by two-sided Kruskal–Wallis test with Dunn’s post-hoc test (a, d, e), or two-sided Student t-test (g). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.