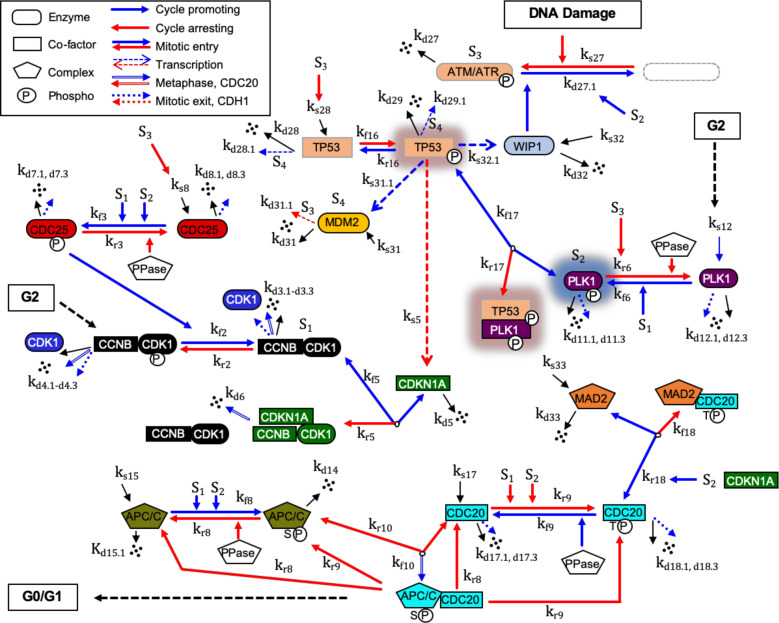
Fig. 1. G2/M DNA damage checkpoint regulatory network.
The G2/M DNA damage biopathway for the reaction network of key proteins is converted into a mathematical structure based on ODEs involving the law of mass conservations and a hybrid framework combining mass action and Michaelis-Menten kinetics. Each factor, as defined in the text and Supplementary information, is identified by a unique color. Gene symbols have been used to be consistent with our previous model of the mitotic cell cycle regulation9. Here, S1 denotes active MPF kinase (CCNB:CDK1), S2 denotes active PLK1 kinase (phosphorylated), S3 denotes activation of ATM or ATR-induced DNA damage response, and S4 denotes activities of the E3 ubiquitin ligase, MDM2. PPase represents generic phosphatases. The model consists of 34 ODEs governing 22 interaction reactions (kfn denotes the forward rate constant and krn denotes the reverse rate constant for reaction n) of 50 key mitotic and DNA damage checkpoint regulation proteins and associated protein complexes and includes 15 synthesis reactions (ks: synthesis rate constant) and 26 reactions of multiple degradations (kdn: self-degradation rate constant, kdn.n: degradation rate constant by other factors including APP/CP:CDC20 and APC/CT:CDH1). Synthesis ks represents either expression of a protein factor or accumulation of a specific post translationally modified factor. The crosstalk components in TP53 and PLK1 pathways are highlighted using background shadows.