Figure 1. Analytic workflow.
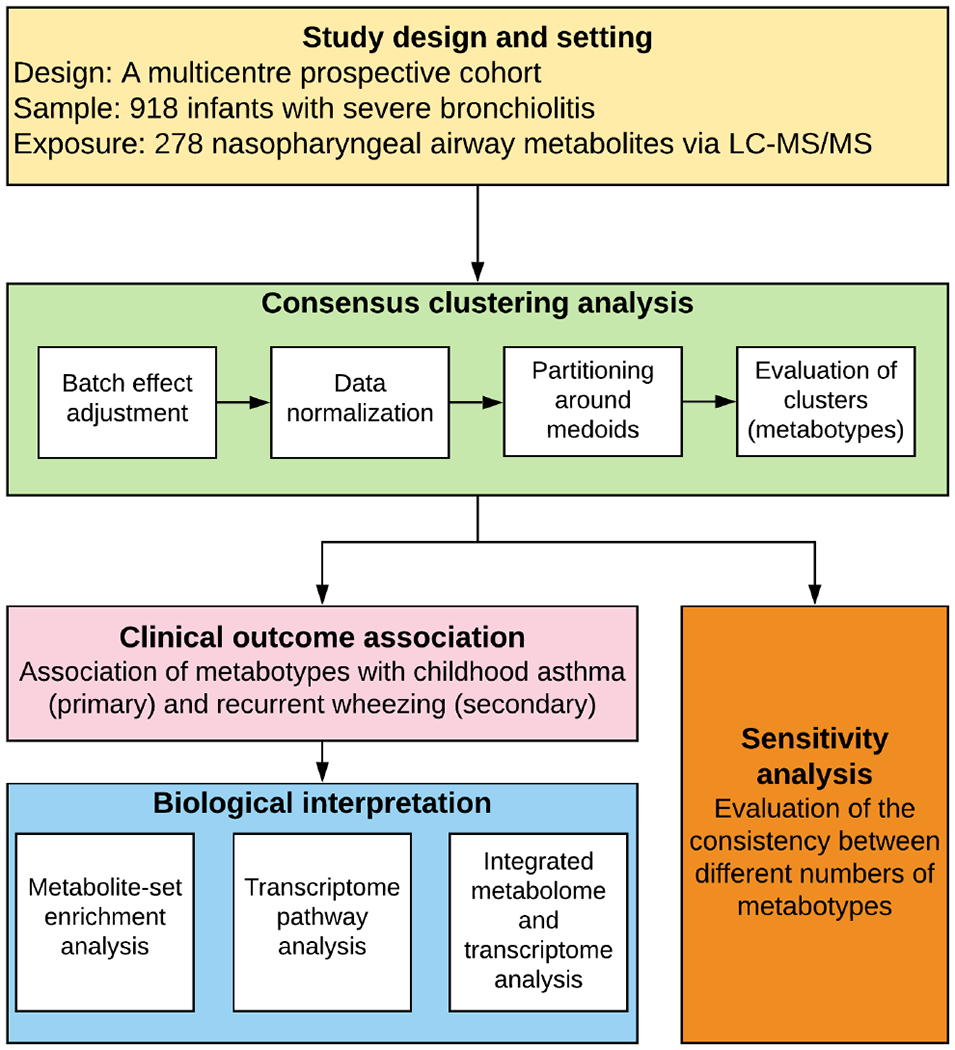
The analytical cohort consists of 918 infants hospitalized for bronchiolitis (severe bronchiolitis) in a multicenter prospective cohort study—the 35th Multicenter Airway Research Collaboration (MARC-35). The nasopharyngeal metabolome data were adjusted for the potential batch effect and normalized by total sum scaling before clustering analysis. To identify metabotypes, the partitioning around medoids algorithm was used in consensus clustering. To choose the optimal number of metabotypes, a combination of the average silhouette score, consensus matrix heatmap, and biological plausibility was used. The between-metabotype differences in the metabolome profile were visualized using a heatmap. The association between the metabotypes and the risk of developing childhood asthma (binary outcome) was estimated by fitting logistic regression models; the rate of developing recurrent wheeze (time-to-event outcome) was estimated by fitting Cox proportional hazards models. To examine the distinct function of each metabotype, 1) metabolite set enrichment analysis examining the metabolome data, 2) gene set enrichment analysis examining the nasopharyngeal transcriptome data, and 3) Wilcoxon pathway enrichment analysis integrating the metabolome and transcriptome data were conducted. In the sensitivity analysis, the concordance between the different numbers of metabotypes was also examined
Abbreviation: LC-MS/MS, liquid chromatography-tandem mass spectrometry
