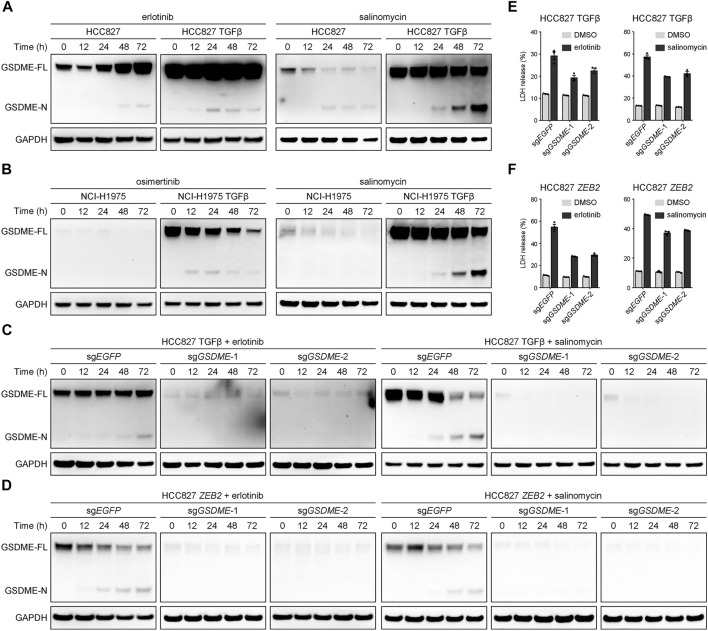
FIGURE 4.
GSDME-dependent pyroptosis in mesenchymal cells upon drug treatment. (A) Parental and TGFβ-stimulated HCC827 cell lines were treated with erlotinib (4 μM) or salinomycin (4 μM) at a time course manner and analyzed by Western blot for GSDME protein cleavage. GSDME-FL, full-length GSDME; GSDME-N, GSDME N-terminal domain. (B) Parental and TGFβ-stimulated NCI-H1975 cell lines were treated with osimertinib (4 μM) or salinomycin (4 μM) at a time course manner and analyzed by Western blot for GSDME protein cleavage. (C) GSDME was knocked out in HCC827 cells using the CRISPR-Cas9 system. Cells were stimulated with TGFβ (5 ng/μl) to induce EMT, treated with erlotinib (4 μM) or salinomycin (4 μM), and analyzed by Western blot for GSDME protein cleavage. (D) GSDME was knocked out in HCC827 cells using the CRISPR-Cas9 system. Cells were transfected with exogenous ZEB2 to induce EMT, treated with erlotinib (4 μM) or salinomycin (4 μM), and analyzed by Western blot for GSDME protein cleavage. (E) GSDME was knocked out in HCC827 cells using the CRISPR-Cas9 system. Cells were stimulated with TGFβ (5 ng/μl) to induce EMT, and LDH release in the presence of DMSO or indicated inhibitors was measured. Data are means ± SEM pooled from three independent experiments. (F) GSDME was knocked out in HCC827 cells using the CRISPR-Cas9 system. Cells were transfected with exogenous ZEB2 to induce EMT, and LDH release in the presence of DMSO or indicated inhibitors was measured. Data are means ± SEM pooled from three independent experiments.