Figure 1.
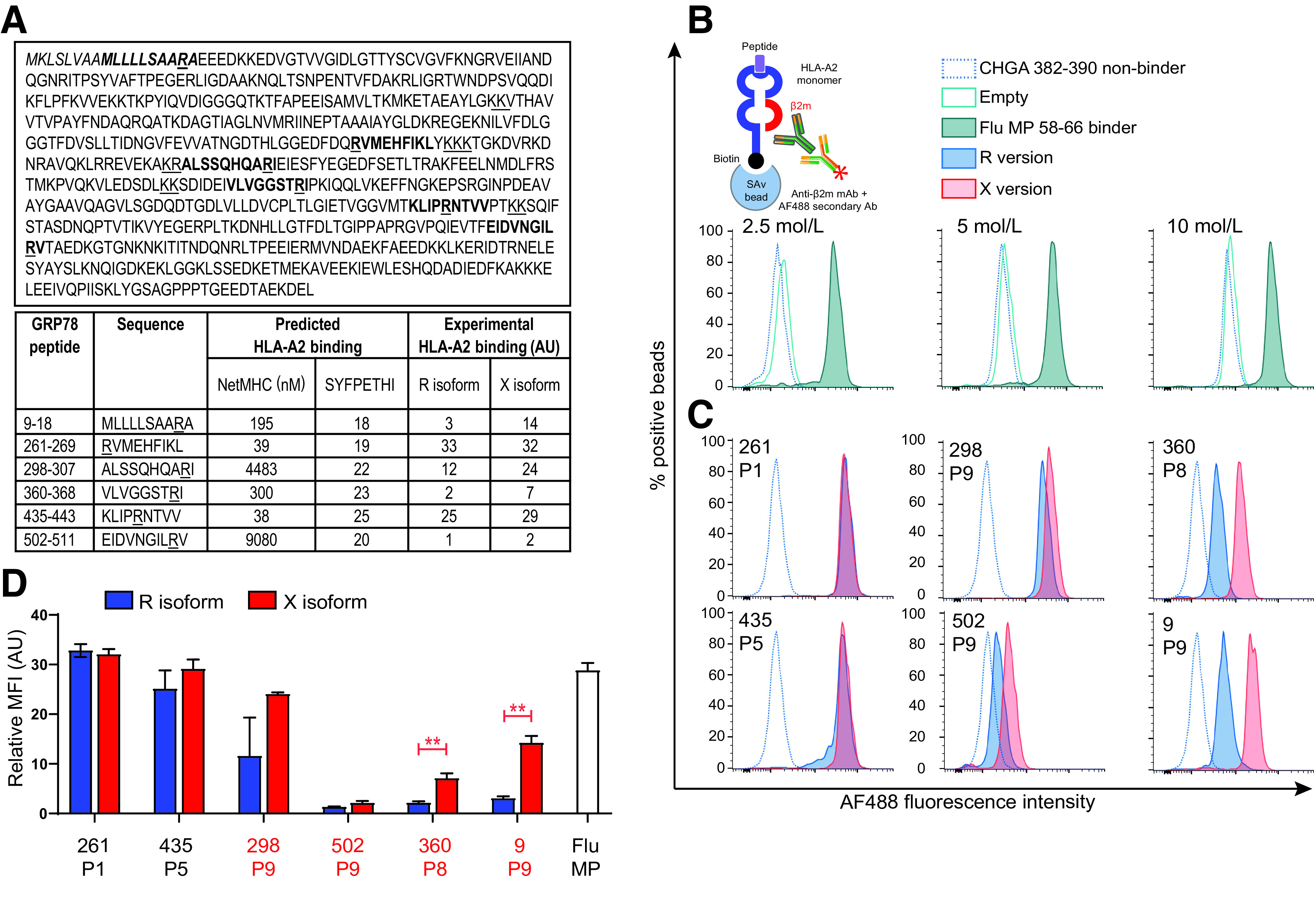
HLA-A2 binding of native (R) and citrullinated (X) GRP78 epitopes. A: Epitope candidates (shown in bold within the GRP78 sequence, UniProt identifier P11021) were selected in silico based on a NetMHC 4.0–predicted KD ≤300 nmol/L or a SYFPETHI score ≥20 (www.cbs.dtu.dk/services/NetMHC and www.syfpeithi.de, respectively). For length variants of the same peptide, the one with the best binding affinity was retained. R residues comprised within the selected candidates and dibasic cleavage motifs are underlined. The GRP78 signal peptide is indicated in italics. The experimental HLA-A2 binding affinity measured for the R and X versions in the experiments shown in subsequent panels is reported in arbitrary units (AU). B: Schematic view and setup of the assay. Biotinylated monomeric HLA-A2 molecules are incubated with peptide and β2m. After capturing on streptavidin-coated beads, an anti-β2m antibody is added, followed by an AF488-labeled secondary antibody. The bead-associated fluorescence is therefore detected only if the test peptide supports the folding of the HLA-A2/β2m complex. Histograms (gated on single AF488+ beads) depict representative flow cytometry staining obtained with increasing concentrations of HLA-A2 molecules folded with the nonbinding CHGA382–390 (HPVGEADYF) and the binding Flu MP58–66 (GILGFVFTL) peptides (negative and positive control, respectively). The 2.5 nmol/L concentration was retained for subsequent experiments based on the best separation of the positive and negative peaks. C: Representative staining of the tested GRP78 peptides in their native (blue) or citrullinated versions (red). The R/X position within the peptide sequence, either outside (P1, P5) or inside (P8, P9) the COOH-terminal region, is indicated in each histogram. D: Relative median fluorescence intensity (MFI) AU values for peptide–HLA-A2 complexes at 2.5 nmol/L, normalized to the ChgA382–390 negative control peptide. Data are expressed as median ± SD of triplicate measurements, and the R/X position is indicated for each peptide. Ab, antibody; mAb, monoclonal antibody. **P ≤ 0.003 by Student t test.
