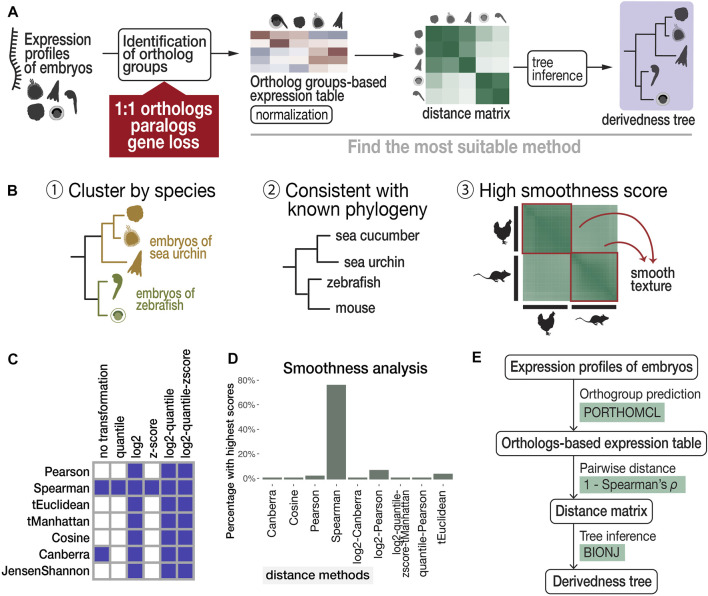
FIGURE 2.
Proposed workflow to estimate phenotypic derivedness of embryos using gene expression profiles and evaluation of methods. (A) Outline of measuring transcriptomic derivedness. Whole embryonic transcriptomes from different species were compared using the expression levels of ortholog groups, which consider not only 1:1 orthologs but also paralogs and genes that are presumably lost in specific lineages. Derivedness index of each embryo was defined as the total branch length from the putative common ancestral node on the inferred tree. (B) Criteria for selecting the suitable method for quantifying derivedness of embryos. These include: [1] clusters samples by species; [2] topology of derivedness tree consistent with known phylogeny estimated from genomic sequences (Kalinka et al., 2010; Hu et al., 2017), with support from biological replicates; [3] Transcriptomic similarities show gradual change along developmental stages. (C) Rank (involved in calculating Spearman distance) and logarithmic normalizations of expression data tend to meet the criteria of clustering stages by species. Shaded boxes represent normalization and distance methods showing this topology in the inferred tree. (D) Spearman distance scores the highest in smoothness analysis. (E) Summary of the most suitable method among the combinations tested.