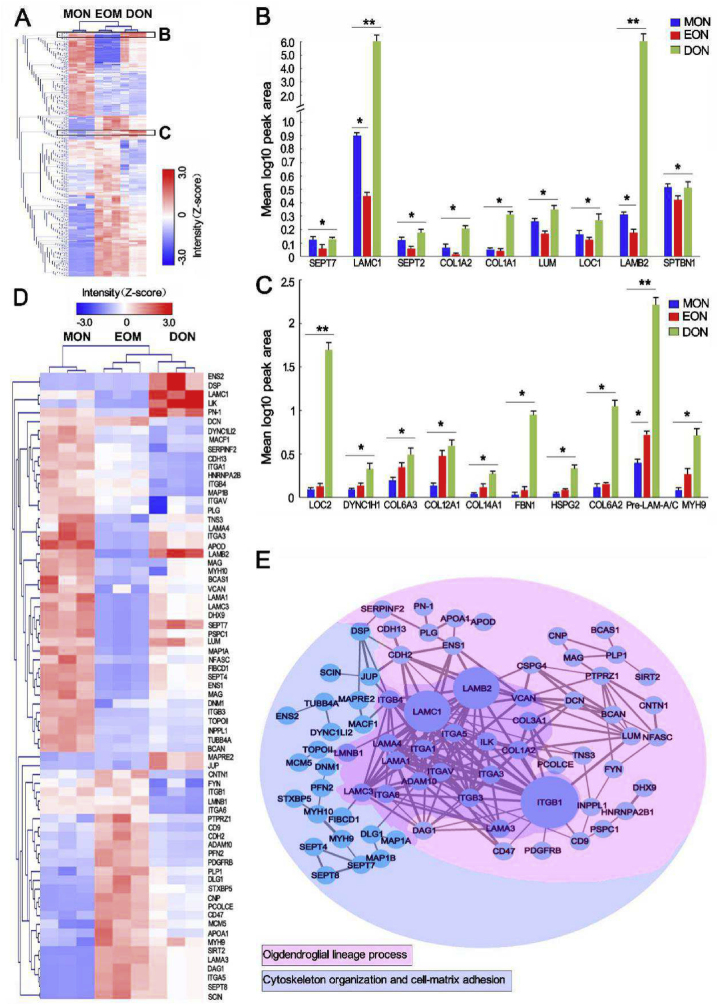
Fig. 2.
Quantitative proteomics analysis of MON, EON and DON. (A) Heatmap representation of proteins that are expressed at significantly different levels (P < 0.05) across the three groups. Z-scored label-free quantification (LFQ) intensities are depicted; red and blue represent increased and decreased values, respectively. (B) and (C) Region of interest (black box in A) containing proteins associated with OL cell maturation (B) and OPC differentiation (C) that were tested for expression. The results were significant across every group, with data input as values to the base 10 to allow for log adjustment (n = 3, **P < 0.0001, *P < 0.001). (D) and (E) Heatmap and protein–protein interaction network constructed from 71 significantly differentially expressed proteins related to lineage development, cytoskeletal organization, and cell-matrix adhesion of OL cells. Z-scored LFQ intensities (D) for which red and blue represent increased and decreased values, respectively. Protein–protein interaction network analysis (E) by strings (confidence cutoff: 0.7). The thickness of the line represents the direct correlation strength of the two proteins, and the size of the nodes (circles) represents the magnitude of the protein effect (networks are based on the string database, https://string-db.org/). The proteins are sorted by color into the two different categories.