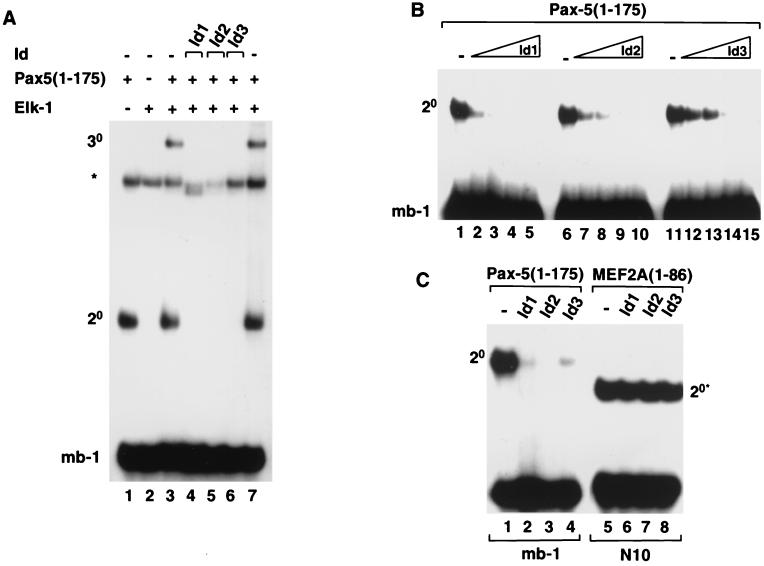
FIG. 2.
Id1, Id2, and Id3 inhibit Pax-5-mediated complex formation on the mb-1 promoter. Gel retardation analysis of complex formation by Pax-5 on the mb-1 promoter in the presence of Elk-1 and/or Id1, Id2, or Id3. The locations of binary Pax-5–DNA (2°) and ternary Pax-5–Elk-1–DNA (3°) complexes are indicated. The asterisk indicates the position of a complex arising from the reticulocyte lysate. (A) Ternary complex formation by in vitro-translated Pax-5(1-175) and full-length recombinant Elk-1 and the mb-1 site in the absence (lanes 3 and 7) or presence of equimolar amounts (with respect to each Id protein) of each of the in vitro-translated Id proteins, Id1 (lane 4), Id2 (lane 5), and Id3 (lane 6). (B) Binary complex formation by Pax-5(1-175) in the presence of increasing amounts of in vitro-translated Id1 (lanes 1 to 5), Id2 (lanes 6 to 10), and Id3 (lanes 11 to 15). The relative molar ratios of Id's added (0, 1, 2, 4, and 8, respectively) are shown schematically above each set of lanes. (C) Id proteins do not inhibit DNA binding by MEF2A. Binary complex formation by equimolar amounts of in vitro-translated Pax-5(1-175) (lanes 1 to 4) and MEF2A(1-86) (lanes 5 to 8) and the mb-1 and N10 sites, respectively, in the absence (lanes 1 and 5) or presence of equimolar amounts (with respect to each Id protein) of each of the in vitro-translated Id proteins, Id1 (lanes 2 and 6), Id2 (lanes 3 and 7), and Id3 (lane 4 and 8). For all reactions, the volumes of reticulocyte lysate added to each lane were equalized by adding appropriate volumes of unprogrammed lysate.