Abstract
Objective
Multimodal automated phenotyping (MAP) is a scalable, high-throughput phenotyping method, developed using electronic health record (EHR) data from an adult population. We tested transportability of MAP to a pediatric population.
Materials and Methods
Without additional feature engineering or supervised training, we applied MAP to a pediatric population enrolled in a biobank and evaluated performance against physician-reviewed medical records. We also compared performance of MAP at the pediatric institution and the original adult institution where MAP was developed, including for 6 phenotypes validated at both institutions against physician-reviewed medical records.
Results
MAP performed equally well in the pediatric setting (average AUC 0.98) as it did at the general adult hospital system (average AUC 0.96). MAP’s performance in the pediatric sample was similar across the 6 specific phenotypes also validated against gold-standard labels in the adult biobank.
Conclusions
MAP is highly transportable across diverse populations and has potential for wide-scale use.
Keywords: phenotype; electronic health records; data mining; biobank, high-throughput
INTRODUCTION
For next generation biobanks enrolling widely across health states and conditions,1–5 methods for high-throughput genomic sequencing are better established and more reliable than methods for ascertaining accurate phenotypes from electronic health record (EHR) data. Computable phenotypes, once developed for a particular condition, can reduce the need to review charts manually to assign a phenotype to each subject.6–8 However, each computable phenotype typically requires a separate effort for feature selection by domain experts and extensive chart review for algorithm training and validation.6,7 In contrast, high-throughput phenotyping algorithms are engineered to require minimal human input and annotation.9 To advance genomic and biobanking research in children, it would be cost- and time-efficient to reuse phenotyping algorithms developed in adults. Algorithms often lose recall and precision when transported from 1 population to another. To date, most studies of transportability of computable phenotype algorithms have focused on rules-based algorithms for identifying specific disease phenotypes; few machine learning-based phenotype algorithms have been validated for transportability.6,8,10,11
We assess transportability of a high-throughput computable phenotyping pipeline from an adult to a pediatric setting. Multimodal automated phenotyping (MAP) is a scalable method for unsupervised phenotyping that can classify millions of subjects across approximately 1800 phenotypes.9 MAP was developed using adult patient EHR data from the Partners Biobank; we applied MAP to EHR data from the PrecisionLink Biobank at Boston Children’s Hospital.2
MATERIALS AND METHODS
Subject selection
Boston Children’s Hospital (BCH) is a freestanding children’s hospital that cares for some adult patients with pediatric conditions (eg, adult congenital heart disease [CHD]). Subject enrollment for the PrecisionLink Biobank at BCH has been described previously2 and is further detailed in the Supplementary Appendix. Written electronic informed consent was obtained at the time of biobank enrollment.2 The BCH Institutional Review Board granted approval—with waiver of informed consent for review of EHR data—for this study.
MAP application
Inputs for MAP included diagnostic codes (grouped into phecodes10,12,13) and clinical note text for all subjects enrolled in the biobank. MAP was applied as previously described9 and as detailed in the Supplementary Appendix available online. The count of all International Classification of Diseases (ICD) codes corresponding to a given phecode (Table 1) was used as the main diagnostic code feature for MAP. To create the main clinical text (referred to subsequently as natural language processing [NLP]) feature for MAP, clinical notes with at least 500 characters were processed using Narrative Information Linear Extraction (NILE) to extract nonnegated concept unique identifiers (CUIs). NILE uses a modified prefix-tree search for named entity recognition and rule-based finite state machines for semantic analyses.14 The extracted CUIs were matched to an automatically curated custom dictionary (Supplementary Methods and Supplementary Figure 1) for each phecode, and only matching CUIs were counted (Table 1 and Supplementary Table 1).
Table 1.
Phenotypes used in the study. Additional concept unique identifiers (CUIs) for each phenotype are listed in Supplementary Table 1. The last column indicates whether validation was performed against unlabeled subjects in the biobank whose charts were manually reviewed or against a registry cohort
| Phenotype | Phecode | CUIs a (Primary and Secondary) | Type of Validation Data |
|---|---|---|---|
| Asthma | 495. | C0004096, C0038218 | Manually reviewed charts |
| Crohn disease | 555.1 | C0010346, C0156147 | Manually reviewed charts |
| Ulcerative colitis | 555.2 | C0009324, C2937222 | Manually reviewed charts |
| Cardiomyopathy | 425. | C0878544, C0007194 | Manually reviewed charts |
| Congestive heart failure; nonhypertensive | 428. | C0018802, C0018801 | Manually reviewed charts |
| Epilepsy, recurrent seizures, convulsions | 345. |
C0014544, C0009951 |
Manually reviewed charts |
| Juvenile rheumatoid arthritis | 714.2 | C0553662, C0157917 | Manually reviewed charts |
| Chronic pulmonary heart disease | 415.2 | C0152171, C0238074 | Manually reviewed charts |
| Type 1 diabetes | 250.1 | C0011854, C0375114 | Manually reviewed charts |
| Cardiac congenital anomalies | 747.1 | C0041207, C0018818 | Registry cohort & Manually reviewed charts |
| Inflammatory bowel disease | 555. |
C0010346, C0009324 |
Registry cohort |
Unified Medical Language System Release 2012AA.
For each phenotype, the candidate cohort consisted of subjects with at least 1 ICD code for the phenotype of interest. MAP models included the total number of ICD codes for each subject as a proxy for healthcare utilization in order to adjust for the noise incurred by unbalanced healthcare utilization.15 MAP uses an ensemble mixture modelling strategy9 on “filter positive” subjects while the risk probability for the “filter negative” subjects is set as 0. We define “filter positive” subjects as those with ICD code and CUI counts both greater than 0. For each phenotype, the probabilities predicted by MAP are cut to create a binary yes/no classification for that phenotype at a cutoff point such that the prevalence of the condition equals the prevalence estimated from gold-standard labels, as described below.
Predicted phenotype evaluation
We first validated the predicted phenotypes against physician-determined diagnoses of 1 of 10 phenotypes (Table 1). Further details are provided in the Supplementary Appendix. We also evaluated the performance of MAP in the PrecisionLink Biobank using subjects enrolled in 2 investigator-driven, disease-specific registries (CHD and inflammatory bowel disease [IBD]). All subjects in each registry have the disease of interest, as determined by clinicians in that field. We excluded registries that were not disease- or condition-specific (eg, “Pulmonary Biobanking Initiative”) or those for a phenotype for which a phecode does not exist (Supplementary Table 2). We excluded registries with fewer than 200 enrolled subjects to ensure the stability of the model evaluation method. We evaluated model performance using subjects with only positive labels and unlabeled subjects with at least 1 relevant ICD code using the method of Zhang et al.16 To enable their method, we modeled the true disease status versus the logarithm of ICD code count and logit of the MAP-predicted probability using piecewise linear spline models, with the number of knots equaling the size of registry data to the one-fifth power. We compared AUCs estimated from positive-only labels with the AUCs from gold-standard chart review performed for patients with CHD, Crohn’s disease, and ulcerative colitis.
Six phenotypes were evaluated against gold-standard labels at both the adult institution at which MAP was originally developed and, in the current study, at BCH. We compared performance of MAP versus ICD codes alone for these conditions at the 2 institutions.
To compare the populations at the 2 institutions, we compared demographic data between institutions as well as histograms of counts of ICD and NLP features at the 2 institutions. We also assessed differences in NLP features between MAP-predicted cases and controls using the same approach. We estimated 95% confidence intervals for the evaluation parameters using the standard bootstrap with 500 replications. All analyses were performed using R version 3.5.1 (R Foundation for Statistical Computing). MAP is available from https://cran.r-project.org/package=MAP
RESULTS
Demographics of subjects in the PrecisionLink Biobank (N = 14 303) and the Partners Biobank, on which MAP was initially evaluated, are shown in Supplementary Table 3. Distribution of ICD counts, length of stay, and CUI counts differed between cohorts both overall and for specific phenotypes (Supplementary Figures 2–4). As expected, the most prevalent phecodes at each institution showed little overlap (Supplementary Table 4).
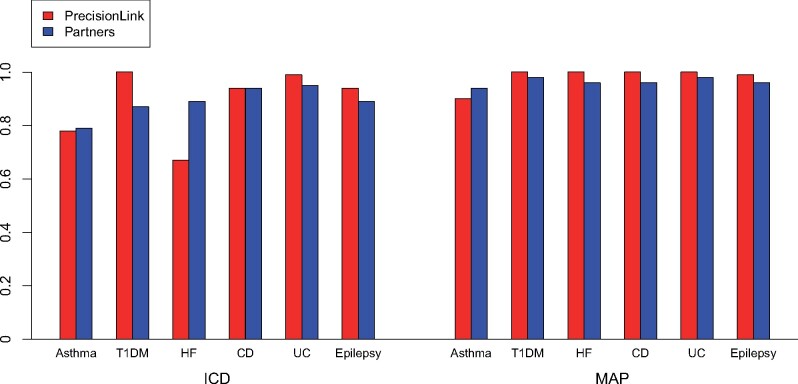
The classification performance of MAP was superior to phenotyping using only ICD codes for all phenotypes except cardiomyopathy (Table 2). Using NLP features alone led to a substantial false positive rate (FPR), reflecting overlap in the distribution of CUI counts between subjects whom MAP predicted to have the phenotype and those predicted not to have the phenotype (Supplementary Figure 5). Overall, MAP performed equally well at the children’s hospital (average area under the receiver operating characteristic curve [AUC] 0.98) as it did at the general adult hospital system (average AUC 0.96). MAP had AUC of 100% for 3 of the 10 phenotypes (Table 2). MAP’s performance on the pediatric sample was similar across the 6 phenotypes also validated against gold-standard labels in the adult biobank9 (Figure 1). In contrast, ICD code-based phenotyping performance had more variability between institutions (Figure 1).
Table 2.
Comparison of computable phenotype algorithm performance using diagnostic codes (ICD), concept unique identifiers (NLP), and multimodal automated phenotyping (MAP). FPR and AUC are shown with 95% confidence intervals. Cohorts marked (RC) are registry cohorts. The remaining cohorts were evaluated using Biobank subjects without labels for a random selection of whom we reviewed medical records.
| Disease | Number Validated | Number Positive | FPR |
AUC |
||||
|---|---|---|---|---|---|---|---|---|
| ICD | NLP | MAP | ICD | NLP | MAP | |||
| Asthma | 20 | 10 | 0.5 (0.21, 0.79) | 0.5 (0.22, 0.78) | 0.2 (0, 0.45) | 0.78 (0.57, 0.99) | 0.67 (0.41, 0.94) | 0.9 (0.76, 1) |
| CD | 20 | 17 | 0.33 (0, 0.88) | 0.33 (0, 0.93) | 0 (0, 0) | 0.94 (0.84, 1) | 0.96 (0.86, 1) | 1 (1, 1) |
| UC | 20 | 15 | 0.4 (0, 0.86) | 0.2 (0, 0.6) | 0 (0, 0) | 0.99 (0.96, 1) | 0.97 (0.91, 1) | 1 (1, 1) |
| CM | 20 | 7 | 0 (0, 0) | 0 (0, 0) | 0.08 (0, 0.22) | 1 (1, 1) | 1 (1, 1) | 0.99 (0.96, 1) |
| HF | 20 | 5 | 0.33 (0.08, 0.59) | 0.13 (0, 0.30) | 0.07 (0, 0.20) | 0.67 (0.42, 0.93) | 0.95 (0.85, 1) | 0.99 (0.94, 1) |
| Epilepsy | 20 | 9 | 0.09 (0, 0.27) | 0.18 (0, 0.41) | 0.09 (0, 0.24) | 0.94 (0.83, 1) | 0.92 (0.78, 1) | 0.99 (0.95, 1) |
| JIA | 20 | 12 | 0.12 (0, 0.35) | 0.12 (0, 0.42) | 0.12 (0, 0.38) | 0.94 (0.84, 1) | 0.89 (0.7, 1) | 0.98 (0.91, 1) |
| PH | 91 | 66 | 0.24 (0.07, 0.41) | 0.24 (0.07, 0.41) | 0.08 (0, 0.18) | 0.94 (0.89, 0.99) | 0.95 (0.91, 0.99) | 0.98 (0.96, 1) |
| T1DM | 20 | 19 | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) | 1 (1, 1) | 1 (1, 1) | 1 (1, 1) |
| CHD | 20 | 15 | 0.4 (0, 0.85) | 0.8 (0.42, 1) | 0.2 (0, 0.63) | 0.8 (0.47, 1) | 0.63 (0.32, 0.95) | 0.93 (0.82, 1) |
| CHD (RC) | 1297 | 0.2 (0.17, 0.24) | 0.19 (0.05, 0.32) | 0.06 (0.02, 0.09) | 0.88 (0.85, 0.91) | 0.85 (0.77, 0.93) | 0.95 (0.93, 0.97) | |
| IBD (RC) | 255 | 0.34 (0.14, 0.54) | 0.21 (0, 0.48) | 0.18 (0, 0.38) | 0.84 (0.69, 0.99) | 0.94 (0.84, 1) | 0.95 (0.85, 1) | |
Abbreviations: AUC, area under the receiver operating characteristic curve; CD, Crohn’s disease; CHD, congenital heart disease; CM, cardiomyopathy; FPR, false positive rate; HF, heart failure; IBD, inflammatory bowel disease; ICD, International Classification of Diseases; JIA, juvenile idiopathic arthritis; MAP, multimodal automated phenotyping; NLP, natural language processing; PH, pulmonary hypertension; T1DM, type 1 diabetes mellitus; UC, ulcerative colitis.
Figure 1.
Comparison of the area under the receiver operating characteristic curve of phenotyping methods for 6 diseases investigated at both Boston Children’s Hospital PrecisionLink Biobank and the Partners Biobank.
Abbreviations: CD, Crohn’s disease; HF, heart failure; ICD, International Classification of Diseases; MAP, multimodal automated phenotyping; T1DM, type 1 diabetes mellitus; UC, ulcerative colitis.
To explore whether we could reduce the burden of manual chart review usually required to test transportability of an algorithm, we estimated MAP’s performance based on existing positive labels from patients enrolled into disease-specific registries within the pediatric biobank. Because these registries enrolled only subjects with a specified condition and not those without that condition, these registry cohorts do not have “true negative” labeled subjects. MAP’s performance was similar when estimated using positive labels from the registry cohorts (AUC = 0.95 for CHD and IBD) and by chart review (AUC = 0.93 for CHD and AUC = 1 for both IBD phenotypes) (Table 2). The MAP algorithm also identified nearly 50% more subjects with CHD (1943 PrecisionLink subjects predicted by MAP vs 1297 subjects enrolled in the registry) and 3 times the number with IBD (1986 subjects predicted vs 487 enrolled in the registry) as were enrolled in either registry, with an estimated false positive rate of less than 20% for all phenotypes.
DISCUSSION
MAP, a high-throughput algorithm for computable phenotyping using EHR data, performed equally well at a freestanding children’s hospital as it did at the adult institution at which it was first developed. MAP’s performance is superior to phenotyping using only ICD codes because of incorporation of knowledge extracted from the clinical narrative into the predictive algorithm. While the benefit of unstructured data for computable phenotyping has been widely demonstrated,8,17 we hypothesize that MAP’s transportability stems partly from use of clinician descriptions of clinical conditions in narrative notes, reducing the algorithm’s dependence on laboratory values, which may vary by assay and patient demographics.18
MAP was previously shown to be highly scalable.9 The current study shows that this scalability extends across practice settings and patient populations. Even without manual feature engineering or supervised training, MAP’s performance at a pediatric site was as good as or better than its performance at the adult institution at which it was developed. The only human input needed to transport the algorithm was to create gold-standard labels to evaluate algorithm performance. Furthermore, as previously shown with simulated data,16 we demonstrate with real-world data that even the effort for this chart review step can be reduced when registry cohorts are available.
It is notable that while many of the conditions used to test MAP’s performance are conditions with little clinical overlap, others, such as heart failure and cardiomyopathy or Crohn disease and ulcerative colitis, have more overlapping features. For all these conditions except cardiomyopathy, ICD codes had a higher FPR, whereas the FPR for MAP remained at or near zero. MAP’s additional strength for these conditions likely stems from combining information from clinical notes and diagnostic codes into the phenotyping algorithm. In contrast, the FPR for asthma, though lower with MAP than with ICD codes alone, remained 20%. This variation may come from less specific descriptions of asthma in the pediatric setting, where distinguishing early childhood wheezing from true asthma can be challenging.19 Supporting this hypothesis, the number of CUIs for the asthma phenotype varied less between cases and controls than for other phenotypes, such as type 1 diabetes (Supplementary Figure 5).
Transportability is an essential component of scalable machine learning algorithms,20,21 as it provides a means for collaborating across institutions in large-scale population-based research.6 Many prior phenotyping algorithms have shown a decrease in performance when transported to other settings.6,22 Although retraining models locally can improve performance,6 such efforts require additional, expensive human annotator effort. Thus, 1 of the benefits we demonstrate for MAP is its ability to maintain performance across settings without additional human annotation.
This study has several limitations. Repetition of text copied from prior notes is common,23 and MAP does not account for such repetition. MAP does control for the total number of ICD codes for each subject, which may help account for some subjects having more encounters with the healthcare system and thus having notes that are more likely to have copied text. Future iterations to the algorithm will explore the added value of accounting for the percent of text in a note that appears to be copied from prior notes. Many of our phenotype predictions were evaluated based on chart review of only 20 subjects per phenotype. While this results in wide confidence intervals for performance for those phenotypes in which performance was less than perfect, average AUC across all phenotypes is based on the review of 271 charts. In future work, we plan to use a semi-supervised learning approach to enhance the statistical power of MAP’s evaluation by combining gold-standard labels with large amounts of unlabeled data.24 Finally, because MAP’s performance depends on conditions being identifiable using CUIs and diagnostic codes, granular endotypes or rare conditions may be less well predicted and require targeted, lower-throughput approaches.
MAP thus represents a transportable, scalable approach for high-throughput phenotyping, enabling approaches such as phenome-wide association studies (PheWAS), where the goal is to create hundreds or thousands of phenotypes simultaneously across a biobank cohort.12 Famously, in a battle of search engine giants, Google outlasted Yahoo by automating its processes while Yahoo relied heavily on manual curation. Our study represents 1 step toward a more Google-like approach for high-throughput biomedical informatics research.25 MAP’s high performance with commonly available input data makes it ideal for use in cross-institutional studies.
FUNDING
This work was supported by the following grants from the National Institutes of Health: NICHD K12HD047349, NHLBI L40HL133929, and NCATS U01TR002623. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
KDM obtained funding. AG, ML, TC, and KDM wrote the manuscript. AG, TC, and KDM designed the research. AG, ML, VAP, and TC performed the research. AG, ML, TC, and KDM analyzed the data. PA contributed new analytical tools. VAP and PA critically revised the manuscript for important intellectual content. All the authors take responsibility for the final approval of the version to be published and are accountable for all aspects of the work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Information Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Florence T. Bourgeois, MD, MPH, for her assistance with clinical record review.
DATA AVAILABILITY STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no financial or other conflicts of interest.
REFERENCES
- 1. Gutiérrez-Sacristán A, De Niz C, Kothari C, Kong SW, Mandl KD, Avillach P. GenoPheno: cataloging large-scale phenotypic and next-generation sequencing data within human datasets. Brief Bioinform 2021; 22 (1): 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bourgeois FAvillach PKong SW, et al. . Development of the Precision Link Biobank at Boston Children’s Hospital: Challenges and Opportunities. JPM 2017; 7 (4): 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henderson GE, Cadigan RJ, Edwards TP, et al. Characterizing biobank organizations in the US: results from a national survey. Genome Med 2013; 5 (1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaught J, Kelly A, Hewitt R.. A review of international biobanks and networks: success factors and key benchmarks. Biopreserv Biobank 2009; 7 (3): 143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012; 19 (e1): e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geva A, Gronsbell JL, Cai T, et al. A computable phenotype improves cohort ascertainment in a pediatric pulmonary hypertension registry. J Pediatr 2017; 188: 224–31.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26 (11): 1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013; 31 (12): 1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong J, Horwitz MM, Zhou L, et al. Using machine learning to identify health outcomes from electronic health record data. Curr Epidemiol Rep 2018; 5 (4): 331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010; 26 (9): 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform 2019; 7 (4): e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu S, Cai T A. Short Introduction to NILE. CoRR2013; http://arxiv.org/abs/1311.6063.
- 15. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc 2018; 25 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L, Ding X, Ma Y, et al. A maximum likelihood approach to electronic health record phenotyping using positive and unlabeled patients. J Am Med Inform Assoc 2020; 27 (1): 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glicksberg BS, Miotto R, Johnson KW, et al. Automated disease cohort selection using word embeddings from Electronic Health Records. Pac Symp Biocomput 2018; 23: 145–56. [PMC free article] [PubMed] [Google Scholar]
- 18. Sagers L, Melas-Kyriazi L, Patel CJ, et al. Prediction of chronological and biological age from laboratory data. Aging (Albany NY) 2020; 12 (9): 7626–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beigelman A, Bacharier LB.. Management of preschool recurrent wheezing and asthma: a phenotype-based approach. Curr Opin Allergy Clin Immunol 2017; 17 (2): 131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Justice AC, Covinsky KE, Berlin JA.. Assessing the generalizability of prognostic information. Ann Intern Med 1999; 130 (6): 515–24. [DOI] [PubMed] [Google Scholar]
- 21. Weng C, Shah NH, Hripcsak G.. Deep phenotyping: Embracing complexity and temporality-Towards scalability, portability, and interoperability. J Biomed Inform 2020; 105: 103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rasmy L, Wu Y, Wang N, et al. A study of generalizability of recurrent neural network-based predictive models for heart failure onset risk using a large and heterogeneous EHR data set. J Biomed Inform 2018; 84: 11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thornton JD, Schold JD, Venkateshaiah L, et al. Prevalence of copied information by attendings and residents in critical care progress notes. Crit Care Med 2013; 41 (2): 382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gronsbell JL, Cai T.. Semi-supervised approaches to efficient evaluation of model prediction performance. J R Stat Soc B 2018; 80 (3): 579–94. [Google Scholar]
- 25. Nicas J. Google took different approaches than Yahoo. The Wall Street Journal 2016 July 26, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.