Figure 4.
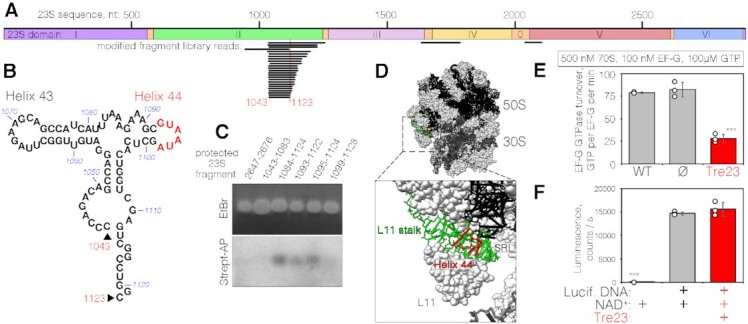
Tre23 ADP-ribosylates helix 44 of 23S rRNA of the GAC center. (A) Mapping of Tre23 ADP-ribosylation by biotinylated fragment enrichment. Sequenced library reads are shown below the diagram representing the architecture of 23S rRNA. The minimal overlapping sequence (nucleotides 1043–1123) is indicated below. (B) Secondary structure of the fragment corresponding to nucleotides 1043–1123. Positions of helices 43 and 44 are indicated, as well as the biotinylated region (red). (C) RNA Protection assay. rRNA-DNA duplexes corresponding to the protected region indicated on top were stained with EtBr (top panel) or detected using streptavidin-AP (bottom panel). (D) Location of the Tre23 ADP-ribosylated 23S region within the 70S ribosome (top, PDB: 6O9J (70)) and a zoom-in on the detected modification position (bottom). The region corresponding to library mapping is shown in green while the modified region, identified by the protection assays, is shown in red. (E) GTPase activity of 100 nM of EF-G was assayed in the presence of 500 nM 70S ribosomes purified from either E. coli MRE600 (WT), E. coli BW25113 cells harboring the empty pNDM220 vector (Ø) or E. coli BW25113 producing Tre23 toxin. Error bars represent standard deviations of the mean of three independent experiments (open circles). *** indicates statistically significant differences between measurements of WT and Tre23-affected ribosomes (unpaired t-test, P <0.0001) or ribosomes purified from cells carrying empty expression vectors and Tre23-affected ribosomes (P = 0.0006). Difference between wild-type and ribosomes from cells carrying empty vector is not statistically significant. (F) Effect of Tre23 on eukaryotic translation in rabbit reticulocyte lysate-based transcription-translation system in presence of DNA encoding the luciferase and purified Tre23 toxin, as indicated. All reactions were supplemented with 0.1 mM NAD+. Luciferase activity is reported in counts per second. Error bars represent standard deviations of the mean of three independent experiments (open circles). *** indicates statistically significant difference between negative control reaction omitting the DNA coding for luciferase and other reactions producing luciferase control in absence or in presence of Tre23 toxin (unpaired t-tests, P < 0.0001). Difference between reactions in absence or in presence of Tre23 toxin is not statistically significant.
