Figure 4.
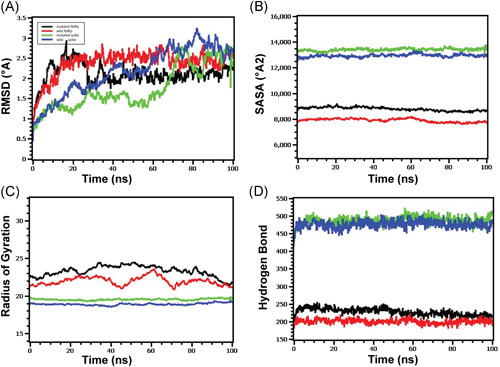
Molecular dynamics of spike‐elastase2 and RdRp‐NSP8 complex (A) Both the wild and mutated spike protein had lower RMSD profile till 60 ns, then it rose and maintained a steady state. Although the spike protein had a higher degree of deviation in the RMSD profile than RdRp, they did not exceed 3.0 Å. The RMSD demonstrated that mutant and wild RdRp protein complex has an initial rise in RMSD profile due to flexibility. Therefore, both RdRp complexes stabilized after 30 ns and maintained a steady peak. The wild‐type RdRp complex had a slightly higher RMSD peak than mutant RdRp, indicating the more flexible nature of the wild‐type. (B) The spike protein complex had a similar SASA profile, did not change its surface volume, and maintained a similar trend during the whole simulation time. The higher deviation of SASA indicates that mutant and wild‐type RdRp had a straight line. Still, the mutant structure had a higher SASA profile, indicating the protein complex had enlarged its surface area. Therefore, the mutation in RdRp protein leads to more surface area expansion than wild types as their average SASA value had a significant difference. (C) Mutated spike exhibits a little more Rg profile than the wild‐type, which correlates with the comparative labile nature of the mutant. The higher level of Rg value defines the loose packaging system and mobile nature of the protein systems. The mutant RdRp had a lower level of fluctuations and maintained its integrity during the whole simulation time. The wild‐type RdRp complexes had higher deviations and more mobility than the mutant complex. (D) Any aberration in hydrogen bond numbers can lead to higher flexibility. Therefore, the mutant and wild spike proteins exhibit the same flexibility in terms of H‐bonding. The mutant RdRp protein had more hydrogen bonding than the wild types, but they did not differentiate too much, and a relatively straight line was observed for the protein. RMSD, root‐mean‐square deviation
