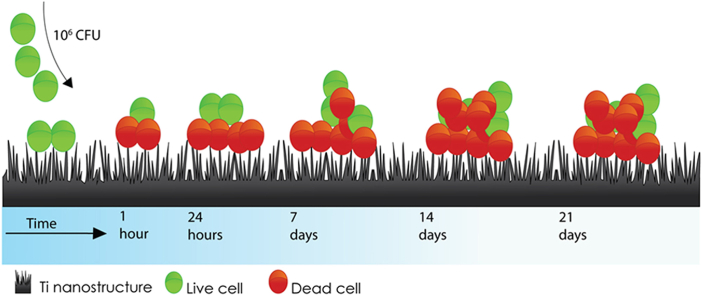
Abstract
The demand for joint replacement and other orthopedic surgeries involving titanium implants is continuously increasing; however, 1%–2% of surgeries result in costly and devastating implant associated infections (IAIs). Pseudomonas aeruginosa and Staphylococcus aureus are two common pathogens known to colonise implants, leading to serious complications. Bioinspired surfaces with spike-like nanotopography have previously been shown to kill bacteria upon contact; however, the longer-term potential of such surfaces to prevent or delay biofilm formation is unclear. Hence, we monitored biofilm formation on control and nanostructured titanium disc surfaces over 21 days following inoculation with Pseudomonas aeruginosa and Staphylococcus aureus. We found a consistent 2-log or higher reduction in live bacteria throughout the time course for both bacteria. The biovolume on nanostructured discs was also significantly lower than control discs at all time points for both bacteria. Analysis of the biovolume revealed that for the nanostructured surface, bacteria was killed not just on the surface, but at locations above the surface. Interestingly, pockets of bacterial regrowth on top of the biomass occurred in both bacterial species, however this was more pronounced for S. aureus cultures after 21 days. We found that the nanostructured surface showed antibacterial properties throughout this longitudinal study. To our knowledge this is the first in vitro study to show reduction in the viability of bacterial colonisation on a nanostructured surface over a clinically relevant time frame, providing potential to reduce the likelihood of implant associated infections.
Abbreviations: IAI, Implant associated infections; HTE, Hydrothermally etched titanium
Keywords: Nanoprotrusions, Nanostructures, Nanospikes, Antibacterial, Biofilm, Implant infection, Biomimetic, Implant associated infections, Orthopedic
Graphical abstract
1. Introduction
Orthopedic implants and other devices play a crucial role in restoring health, treating musculoskeletal disease, and ultimately saving patient lives [1]. Titanium alloy is widely used as a biomaterial to replace hard tissue, due to its mechanical bone-like properties, enhanced corrosion resistance and superior biocompatibility [2]. However, implant associated infection (IAI) occurs in 1%–2% of joint replacement [3,4] and approximately 30% of open fracture reduction cases [5,6], leading to notably increased morbidity, mortality, and treatment cost [7,8]. IAI involves a series of complex interactions between the pathogen, the implant, and the host immune response. In the absence of a foreign body, tissue contamination by opportunistic pathogens is usually spontaneously cleared by host immune defences. However, the presence of an implant triggers a localised tissue response, which includes acute and chronic inflammation, a foreign body reaction, formation of granulation tissue and, finally, fibrous encapsulation. In this early phase, bacteria may take advantage of the immunocompromised region around the implant to adhere, colonise and form a biofilm [9]. Ideally, in a concept termed ‘race to the surface’, host cells outcompete bacteria to colonise the implant surface, leading to favourable tissue integration – where the host immune system is better placed to prevent the onset of IAI [10,11].
Bacteria adhere to implants by a process mediated by hydrophobic, electrostatic and van der Waals interactions, where they colonise the device surface to form a biofilm which can evade the immune response. A biofilm manifests, following a sequence of growth phases to become a multifaceted biological unit that is extremely difficult to treat, becoming resistant to systemic antibiotics [[12], [13], [14]]. An IAI results in an unresolved chronic inflammatory response, leading to tissue destruction and ultimately loosening of the bone-implant interface [15].
Treatment of an IAI usually involves the removal of the implant and associated cement, debridement of all devitalised tissue, and long-term antimicrobial treatment [16]. Sources of bacterial contamination implicated in IAI include the operating room environment, surgical equipment, clothing worn by medical staff, and bacteria on the patient's skin and residing in the patient's body [[17], [18], [19]]. The most common etiologic agents causing IAIs are Staphylococcus aureus (S. aureus), Staphylococcus epidermidis (S. epidermidis) [20], Pseudomonas aeruginosa (P. aeruginosa) [21], and Escherichia coli (E. coli) [22]. These bacteria are normally associated with skin and environmental microbiota but can quickly colonise implants leading to biofilm formation [23]. Four out of five IAIs are caused by Staphylococcus species, with S. aureus and S. epidermidis responsible for two out of three [20]. Drug-eluting coatings have been successfully attached to implants, including silver nanoparticles [[24], [25], [26]], antibacterial peptides [27,28] and antibiotics [29], amongst others [30,31]. However, these finite eluting treatments are generally short-acting and may harm eukaryotic cells, prompting attempts to find safer and longer-lasting, non-eluting solution [32,33]. Following the discovery that mechano-bactericidal nanostructures found on the wing of a dragonfly can kill a variety of bacterial species upon contact [34], it has been shown that titanium surfaces with a specific nanoarchitecture mimicking that of the dragonfly wing can also be highly effective in killing bacteria [[34], [35], [36]]. Mechano-bactericidal activity of nanostructured surfaces is governed by surface profile and is independent of surface chemistry [37]. Nanostructures with a high aspect ratio - with a length many times that of their width, mechanically disrupt the bacterial membrane [38]. Whilst nanostructured surfaces have shown antibacterial efficacy against Gram-positive species, Gram-negative species such as P. aeruginosa and E. coli are particularly vulnerable, due to differences in cell wall architecture [35,39]. Several methods have been developed to fabricate nanostructures on titanium and its alloys, with antibactericidal properties to enhance osteointegration. Some of these methods include; anodization [40], sol-gel [41] and hydrothermal etched titanium (HTE) [42]. HTE stands out as the method of choice to fabricate TiO2 due to; its simplicity, inexpensive, environmentally friendly, and more importantly can generate a variety of nanoscale patterns [42]. Furthermore, TiO2 surface layer exhibits low solubility in water, which prevents substrate metal ions from dissolution, enhanced wear and fatigue resistance [43].
A limitation of previous studies has been the short time frame over which bacteria were cultured on nanostructured surfaces, whereas biofilms are known to evolve over days and indeed weeks [36,39,[44], [45], [46]]. Moreover, reduced early bacterial viability as well as inhibited and/or delayed biofilm formation on the implant surface may promote improved host tissue integration in the early phase, which further reduces the likelihood of an IAI developing [47,48]. The aim of this study was, therefore, to examine the antibacterial efficacy of a nanostructured titanium alloy on two clinically relevant pathogens (S. aureus and P. aeruginosa) over a 21-day period. We hypothesise that, there will be a continuous, long-term disruption of biofilm growth and maturation on the antibacterial nanostructured implant surface, which may translate to implants with reduced occurrence of IAIs.
2. Materials and methods
2.1. Fabrication of antibacterial Ti6Al4V surface
To create the study HTE surface, medical grade Ti6Al4V discs (10 mm diameter, 3 mm thickness) with a mirror polished top surface and a RA of 0.1 μm (Hamagawa Industrial (M)76 SDN BHD, Kedah Malaysia), were hydrothermally etched, using 1 M KOH in a stainless-steel reactor (autoclave; Parr Instrument Company, USA). After the chemical reaction, the vessels were cooled, and samples were cleaned in ultrapure water. After drying for 2 h at 70 °C, discs were heat treated inside a tubular furnace and cooled down overnight. As-received Ti6Al4V discs with a polished top surface were used as the control implant surface (CTL) in biofilm experiments. All samples were cleaned and sterilised at 121 °C for 20 min prior to use in biological experiments.
2.2. Characterisation of nanotopography by scanning electron microscopy (SEM) and atomic force microscopy (AFM)
The morphology of the HTE and CTL surfaces was characterised using scanning electron microscopy (SEM) (Zeiss Merlin FEG-SEM, Zeiss, Jena, Germany) at 2 KV, 4.5 mm working distances with magnifications from 5 K to 50 K. Substrate surfaces were orientated at 45° relative to the horizontal plane in the SEM, where an oblique view reveals additional information about the nanotopography that cannot be assessed from a top-down perspective, such as approximate nanostructure height. Five samples were used to measure height, diameter and spacing between nanostructures. The height of the nanostructures was calculated using the distance between a perpendicular plane and the highest point of each spike, and the diameter was measured at mid-height in parallel orientation with the basal plane (25 spikes measured per sample). The measurements for spike height were corrected for the 45° stage tilt [49]. To determine the spacing between nanostructures, zero-degree tilted SEM images were analysed (n = 5), considering nanostructure tips in a 5 μm2 field. Analysis was performed using ImageJ software v1.53a (NIH, USA). The morphology of the HTE and CTL surfaces was also characterised in air, using atomic force microscopy (AFM). Data for 5 μm2 surface area scans was acquired using a JPK NanoWizard III (JPK BioAFM, Berlin, Germany) with instrument-specific software v5, written in Linux Ubuntu (Canonical Ltd., London, UK). An NT-MDT NSG03 silicon nitride cantilever with a conical tip rated by the manufacturer (Spectrum Instruments Ltd, Moscow, Russia) at a radius less than 10 nm and a half side angle of 18° was used to perform amplitude modulation or tapping mode scans on HTE and CTL surfaces (n = 3). Initial calibration of the cantilever on a glass microscope slide derived a normal spring constant of 1.9 N/m and a deflection sensitivity of 86.8 kHz. Scanning parameters over a scan rate of 0.8 Hz were held constant at a setpoint of 22.8 nm and a drive amplitude of 0.3 V. Roughness values and 3D reconstructions were acquired using Gwyddion data analysis software version 2.54. (http://gwyddion.net/).
2.3. Contact angle analysis
The water contact angle (WCA) of the CTL and HTE samples (n = 3) was measured using a contact angle goniometer model RD-SDM02 (RD Support, Scotland, UK), to determine surface wettability. Contact angles were measured for five randomly placed drops on triplicate samples. Both CTL and HTE samples were placed on a flat surface to receive a 4 μL of Milli-Q water and the WCA was measured within 20 s using the sessile drop analysis.
2.4. Chemical analysis of the surface by X-ray photoelectron spectroscopy (XPS)
XPS was employed to investigate the surface chemistry of the CTL and HTE samples (n = 3). Survey spectra were collected with a pass energy of 160 eV and high-resolution spectra were obtained using a 20-eV pass energy, with a monochromatic Al source run at 15 keV and 15 mA on a Kratos AXIS Ultra DLD spectrometer (Kratos Analytical Ltd, Manchester, UK). Survey spectra were documented over a 0–1100 eV range at 0.5 eV increments. CasaXPS software version 2.3.23 (www.casaxps.com, Teignmouth, UK) was used to process the spectra after curve fitting, and all binding energies were referenced to the carbon peak at 285.0 eV.
2.5. Energy-dispersive X-ray spectroscopy (EDS) for elemental analysis
HTE samples were dry cut with a CBN blade from More Superhard (Zhengzhou, China) at 300 rpm in a Struers Minitome (Willich, Germany). To compare the qualitative elemental analysis between the HTE surface and the subsurface, a Zeiss Merlin FEG-SEM (Zeiss, Jena, Germany) equipped with an EDS detector (Oxford Instruments X-MaxN (20 mm2) EDS-Silicon Drift Detector) was used. Spectra were obtained, using a SE2 detector at 15 kV, 3 nA, working distance 10 mm, analytic mode and 10 K magnification, using AZTEC EDS software version 3.1 (Oxford Instruments, Oxfordshire, UK).
2.6. Short-term cytocompatibility of the HTE nanostructured surface
The ability of RAW 264.7 macrophage like cells (ATCC® TIB-71™, VA, USA) to adhere and proliferate on the HTE surface was assessed by LIVE/DEAD® Viability/Cytotoxicity Kit for mammalian cells (Molecular Probes, Invitrogen, CA, USA), following the manufacturer's protocol. RAW 264.7 macrophage like cells were cultured in Dulbecco's modified Eagle's medium (DMEM, ThermoFisher, MA, USA) supplemented with 10% fetal bovine serum (FCS; ThermoFisher, MA, USA) and 1% penicillin/streptomycin (ThermoFisher, MA, USA). Cells were seeded on HTE and CTL samples at 2.5 × 104 cells per well in a 48-well tissue culture plate and incubated at 37 °C in 95% humidity and 5% CO2. After 48 h culturing on HTE and CTL samples in a 48-well tissue culture plate, the cells were washed gently with phosphate buffered saline (PBS), followed by calcein AM/EthD-1 working solution and incubated at room temperature for 30 min, then imaged by confocal laser scanning microscope (CLSM; Olympus, Tokyo, Japan). Calcein (Ex/Em 495/515 nm) will fluorescently stain live cells green, whereas EthD-1 (Ex/EM 495/635 nm) will stain cells with damaged membranes or dead cells, producing a bright red fluorescence upon binding to nucleic acids. Viability was quantified by counting red and green stained cells using ImageJ software v1.53a (NIH, USA). Cytotoxicity was further assessed using LDH-Glo™ Cytotoxicity Assay (Promega, WI, USA), to detect extracellular lactate dehydrogenase (LDH), a widely used marker in cytotoxicity studies due to its rapid release in tissue culture media upon disruption of the cell membrane [50,51]. Briefly to quantify LDH release, 50 μL of media was added to 50 μL of LDH detection reagent in a 96-well opaque plate, mixed and incubated at room temperature for 30 min then luminescence read on a Synergy HTX multi-mode microplate reader (Biotek, VT, USA). Triplicate readings were normalised to cells grown on tissue cultures plate (TCP). The formula used to calculate percent cytotoxicity was,
2.7. Bacterial culturing and biofilm growth protocols
P. aeruginosa, ATCC 15692 and S. aureus, ATCC 25923 glycerol stocks were plated onto Tryptone Soy Agar (TSA; Oxoid, ThermoFisher, MA, USA) plates and incubated for 18 h at 37 °C. Single colonies of each species were inoculated into Tryptone Soy Broth (TSB; Oxoid, ThermoFisher, MA, USA) and 5% FCS (TSBFCS; Gibco, ThermoFisher, MA, USA), and incubated overnight at 37 °C. The following day bacterial cultures were gram stained to confirm purity and adjusted to 1 × 106 CFU/mL by adjusting to the associated 600 nm (OD600) reading measured on a Nanodrop 2000C (ThermoFisher Scientific, MA, USA). An OD600 of 1 was determined to be approximately 1 × 109 CFU/mL by calibrating against colony forming units (CFU) for P. aeruginosa and S. aureus prior to the commencement of this study. Samples of HTE and CTL were inoculated in a 24-well plate with 1 mL of 1 × 106 CFU/mL of either P. aeruginosa or S. aureus for nine time points (day 1–7, 14 and 21) and incubated with daily replenishment of TSBFCS, at 37 °C in a humid chamber on an orbital shaker (Ratek Instruments, VIC. Australia), on setting 6. At each time point HTE and CTL samples were removed for analysis.
2.8. Quantitative antimicrobial test
The gold standard for enumerating bacteria is the colony forming unit (CFU) method [52]. Briefly, at each time-point (day 1–7, 14 and 21), HTE and CTL discs were transferred into 1 mL PBS and vortexed for 15 s, followed by 2 min sonication then vortexed a further 15 s, and serially diluted. Serial dilutions were plated onto TSA plates and incubated for 18 h at 37 °C. Viable cell counts were performed on the HTE and CTL surfaces by the drop plate method [50]. The following day, colonies were counted, and CFUs determined for P. aeruginosa and S. aureus incubated on HTE and CTL discs [53]. Comparison between viability on the HTE and CTL samples was further used to determine log reduction, log10 NCTL NHTE and percent reduction for each time-point. To validate complete removal of bacteria the HTE and CTL samples were imaged by SEM, post CFU processing.
2.9. Bacteria live/dead assay
Bacterial cells attached to the surface of HTE, and CTL samples were stained by Live/Dead® BacLight™ (Invitrogen, ThermoFisher MA, USA) according to the manufacturer's protocol. Briefly, a mixture containing Syto9 (Ex/Em 480/500 nm) and Propidium Iodide (PI; Ex/Em 490/635 nm) in equal proportions (1.5 μL of each per mL) in PBS was freshly prepared. Syto9 is taken up by all cells and binds with nucleic acids, staining the cell green. Whereas PI is unable to pass through intact plasma membrane and is therefore only taken up by damaged cells allowing detection of both live (green) and compromised cell membrane integrity that is associated with cell death (red). The samples were immersed in the live/dead solution and incubated at room temperature for 15 min in the dark prior to microscopy.
2.10. Confocal laser scanning microscope analysis
Samples were imaged at each time-point (days 1–7, 14 and 21), using three random images per sample. CTL and HTE samples were imaged for viability and biomass analysis, using an Olympus FV3000 CLSM (Olympus, Tokyo, Japan). Single plane micrographs were used to determine viability by counting red and green stained cells using ImageJ software v1.53a (NIH, USA). Z-stack images were acquired for S. aureus and P. aeruginosa on days 7, 14 and 21. The viability and total biomass for the Z-stacks were also examined using Imaris 3D analysis software (Version 9.3.0, Bitplane, Zürich, Switzerland). The excitation/emission maxima of the LIVE/DEAD Bacterial Viability Kit dyes are 480/500 nm for SYTO 9 and 490/635 nm for PI.
2.11. SEM analysis of bacterial morphology
To prepare biological samples for SEM, samples were fixed in 1.25% glutaraldehyde, 4% paraformaldehyde in PBS containing 4% sucrose for 24 h. Next, they were washed in PBS, followed by dehydration in an ascending ethanol series from 50%, 70% and 100% ethanol. Finally, samples were chemically dried using hexamethyldisilane (Sigma-Aldrich, MO, USA), mounted on aluminium stubs, sputter coated with 1 nm platinum, and imaged using a Zeiss Merlin FEG- SEM (Carl Zeiss, Jena, Germany).
2.12. Statistical analysis
Graphical data was represented by mean and standard deviation. All statistical analysis were performed using GraphPad Prism version 9.0.0.0 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). The data was analysed by multiple t-tests. Statistical significance was determined using the Holm-Sidak method, with alpha = 0.05, significance levels were set at p < 0.05. All experimental procedures were performed in biological replicates (n = 3).
3. Results and discussion
3.1. Characterisation of HTE nanostructure surface
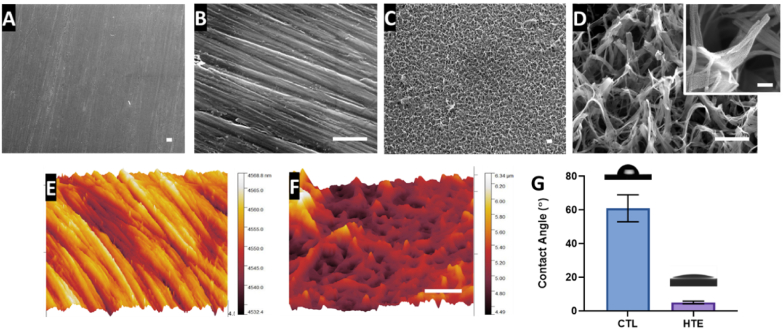
Representative SEM micrographs depicting the nanostructured features of the CTL and HTE samples are shown in Fig. 1A–D at 5, 50 and 200 K magnification, respectively. The CTL surface appeared smooth at lower magnification, however, at high magnification, machining marks and scratches due to the polishing process can be seen. The HTE surface exhibited hierarchal disordered spike-like nanostructures covering the entire surface. The height of the nanostructures was 348 ± 152 nm, and diameter at mid-height was 98 ± 60 nm, signifying a high aspect ratio. The spacing between the individual nanostructures was 437 ± 46 nm. Nanostructures with similar dimensions have previously been shown to be effective at killing both Gram-positive and Gram-negative bacteria [35,38].
Fig. 1.
SEM micrographs of unetched CTL (1A and 1B) showing effects of polishing; and the HTE surface (1C and 1 D) displaying hierarchal nanotopography (scale bar for Fig. 1A–D = 500 nm). The inset image in 1 D shows the morphology of a single nanostructure at 200 K magnification (scale bar = 100 nm). AFM-3D reconstructions showing nanotopological features of a 5 × 5 μm region on CTL (1E) and the superhydrophilic HTE surface (1F) (scale bar = 1 μm). The bar graph represents the water contact angle for CTL and HTE surfaces mean ± SD (n = 3), p = 0.0003. Images above the bar graph demonstrate the wettability by the sessile drop method (1G). Nanostructure measurements, AFM and wettability can be found in supplementary information Table S1.
The AFM image, presented in Fig. 1E, confirmed machining marks due to polishing on the CTL surface. Nanostructures with peaks and valleys can also be seen in the AFM image from the HTE surface (Fig. 1F), however the fine details seen by SEM could not be resolved by AFM due to the well-known tip convolution effect [54]. The AFM images displayed greater roughness on the HTE surface compared to the CTL surface. The root mean square roughness (RMS), arithmetic roughness average (Ra) and surface area (SA) determined from the AFM images for the CTL surface were 5.13 nm, 4.15 nm and 25.14 μm2, respectively, while for the HTE surface these parameters were 236.3 nm, 175.5 nm and 64.64 μm2 respectively. The surface area measured by AFM on the HTE sample increased by 2.6-fold when compared to the CTL. It should be noted, however, that the tip convolution resulting from the cantilever tip side angle and scan velocity reduced the measured surface roughness values, especially towards surface features with comparable magnitude to the tip radius. The water contact angle for the control surface (61 ± 8°) was significantly greater than that of the HTE surface (5 ± 0.8°, p = 0.003, Fig. 1G). The Wenzel [55] and Cassie–Baxter [56] equations state that adding surface roughness will enhance the wettability caused by the chemistry of the surface. It is well known a water contact angle between 0° and 10° is categorised as superhydrophilic [57]. Generally, an increase in hydrophilicity is an imperative characteristic to improve biocompatibility and antibactericidal properties of the HTE surface - where hydrophilic implant surfaces favour cell-implant contact, the initial phases of wound healing and a cascade of events leading to the osteogenesis [[58], [59], [60], [61]].
3.2. Chemical analysis
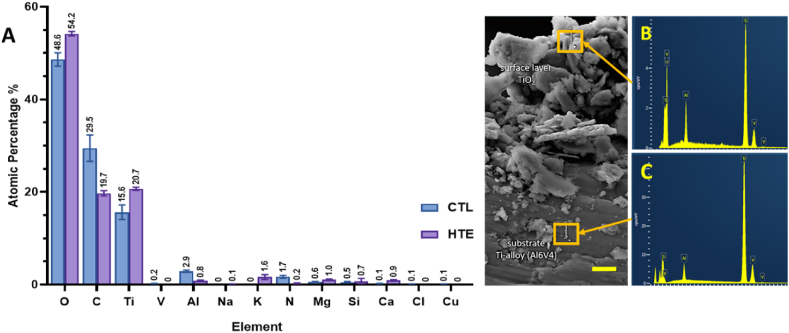
The atomic percentage of elements detected on the CTL and HTE disc surfaces using XPS is shown in Fig. 2A. There was a significant increase in oxygen and titanium atomic concentrations on the HTE surfaces compared to the untreated CTL (5.6%, p = 0.0003 and 5.1%, p = 0.0002 respectively). These results are consistent with formation of a thicker oxide layer occurring during the hydrothermal etching process. The increased thickness of the oxide layer also meant that, with the XPS sampling depth of 10 nm, vanadium and aluminium became barely detectable on HTE surfaces. Hydrothermal etching also reduced the presence of environmental contaminants such as carbon and nitrogen. Furthermore, a small amount of potassium was incorporated within the HTE surface, consistent with the use of KOH as the alkaline etchant. Trace amounts of magnesium, calcium, and silicon, incorporated during polishing and handling of the CTL samples, were detected on both CTL and HTE disc surfaces.
Fig. 2.
Atomic percentages for CTL and HTE samples quantified by XPS, mean ± SD (n = 3). p-values for all elements detected can be found in supplementary information in Table S2 (2A). EDS spectra measured on HTE surface (2B) and substrate (2C) (scale bar = 5 μm).
Using EDS (Fig. 2B and C), we confirmed the increase in oxygen content for the HTE surface, consistent with hydrothermal oxidation of the surface layer occurring during hydrothermal etching discussed above [42]. The three elements used in the manufacture of the Ti-alloy (i.e., Ti, Al, V) were also detectable in the EDS spectra.
3.3. In vitro cytotoxicity of raw 264.7 cells to HTE treated surface
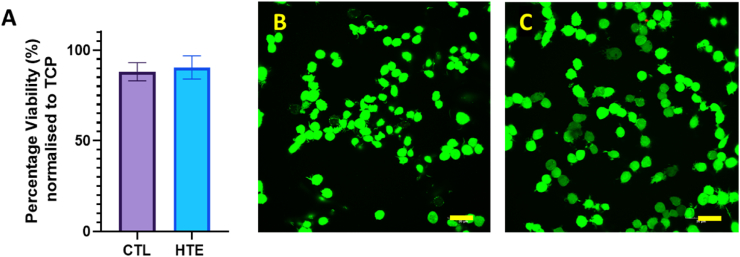
After biomaterial implantation, macrophages are one of the earliest host-cell responders [62]. We therefore used a RAW 264.7 macrophage-like, Abelson leukemia virus-transformed cell line derived from mice, to observe any short-term cytocompatibility to the HTE surface. Briefly, the viability of RAW 264.7 cells seeded onto CTL and HTE discs are shown in Fig. 3A. After 48 h incubation, the percent viability for CTL and HTE discs was 95.8 ± 2.6% and 94.0 ± 4.2%, respectively (p = 0.57). Furthermore, after fluorescent staining with LIVE/DEAD® Viability/Cytotoxicity Kit, no red stained cells (indicating dead cells) were detected on the CTL or the HTE samples. Cells on both CTL and HTE samples appeared healthy and had comparable morphology. The cells were approximately 60% confluent after 48 h on both CTL (Fig. 3B) and HTE samples (Fig. 3C).
Fig. 3.
Viability of RAW 264.7 macrophage-like cells determined by LDH-Glo™ Cytotoxicity Assay, normalised to RAW 264.7 macrophages grown directly on TCP. The mean ± SD percentage viability for RAW 264.7 macrophages incubated on CTL samples was 95.8 ± 2.6% and HTE samples was 94.0 ± 4.2% (mean ± SD), p = 0.57, n = 3 (3A). The confocal images of RAW 264.7 macrophages incubated on the CTL (3B) and HTE (3C) for 48 h, stained with LIVE/DEAD™ Viability/Cytotoxicity Kit, for mammalian cells (scale bars = 30 μm).
3.4. Bacterial viability analysis
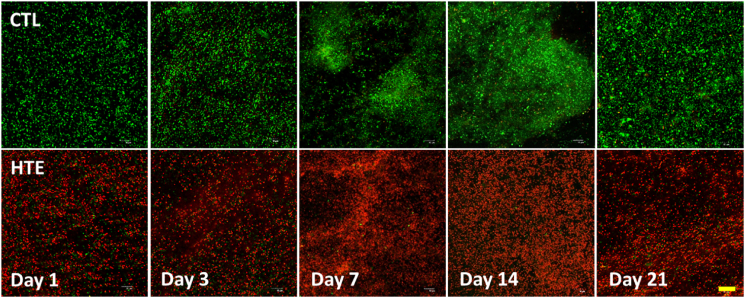
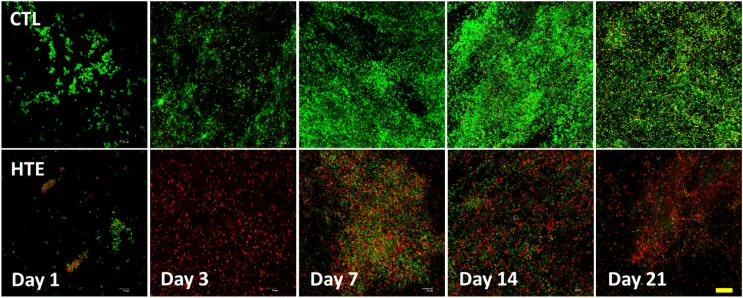
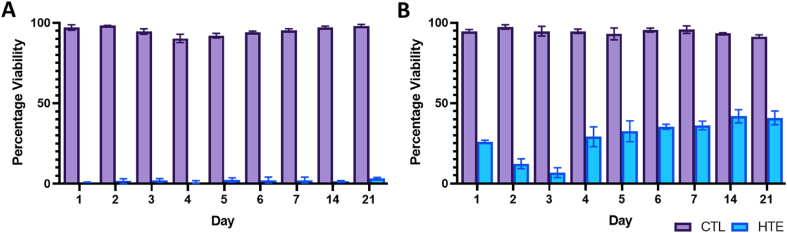
To assess the viability of bacterial populations on the CTL and HTE surfaces, we used LIVE/DEAD® BacLight™ Bacterial Viability Kit [63]. Representative live/dead images of P. aeruginosa and S. aureus at days 1, 3, 7, 14 and 21 are shown in Fig. 4, Fig. 5, respectively. When P. aeruginosa was cultured on HTE surfaces, the bacterial viability was only 0.97 ± 0.06% after 24 h. Longer incubation times resulted in a marginally higher mean viability, which never exceeded 4%. (Fig. 6A). For S. aureus, there was an initial cell viability of 26.0 ± 0.9% after 24 h incubation. On day 3, S. aureus viability was at its lowest (6.8 ± 3.1%), with viability increasing to 36.2 ± 2.7% by day 7, and then 40.9 ± 4.3%, by day 21 (Fig. 6B). In contrast, the viability for S. aureus and P. aeruginosa did not drop below 90% throughout the 21-day study on the CTL surface (Fig. 6A and B).
Fig. 4.
Confocal microscopy images of P. aeruginosa after staining with LIVE/DEAD® BacLight™ Bacterial Viability Kit incubated on CTL and HTE surfaces at five representative time points over 21-days. Live cells stain fluorescent green, whereas bacteria with damaged membranes stain fluorescent red. Top panels represent as-received CTL, and the bottom panels represent the matching time point HTE samples incubated with P. aeruginosa (n = 3). Scale bar = 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Confocal microscopy images of S. aureus after staining with LIVE/DEAD® BacLight™ Bacterial Viability Kit incubated on CTL and HTE at five representative time points over 21 days. Live cells stain fluorescent green, whereas bacteria with damaged membranes stain fluorescent red. The top panels represent as-received CTL, and the bottom panels represent the matching time point HTE samples incubated with S. aureus (n = 3). Scale bar = 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Viability of P. aeruginosa (6A) and S. aureus (6B) determined from the live/dead staining over 21 days on CTL and HTE surfaces, mean ± SD (n = 3). To compare CTL and HTE surfaces, multiple comparisons using the Holm-Sidak method revealed a significance of p < 0.0001 at all-time points. Table of p-values can be found in the supplementary information (Table S3 and S4 respectively).
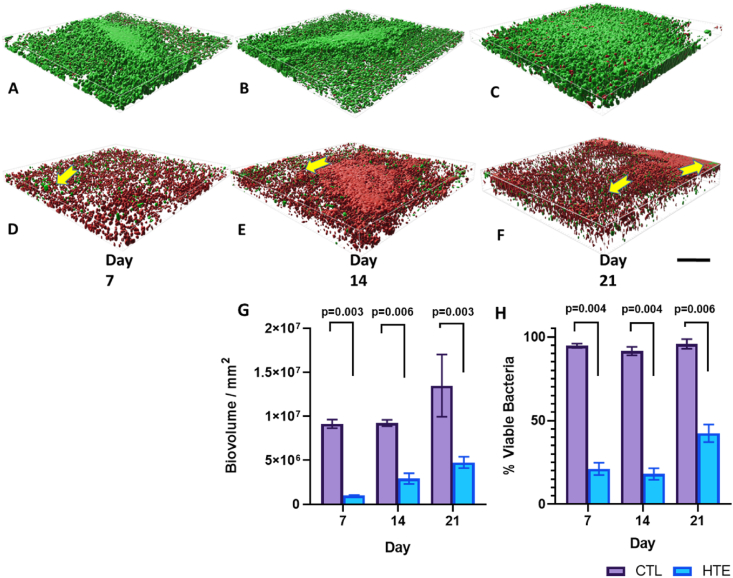
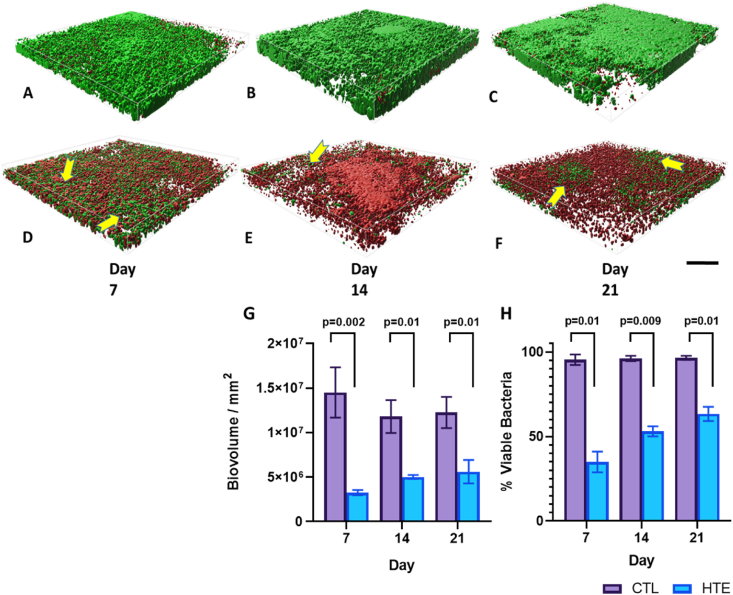
To analyse the biovolume on the CTL and HTE surfaces, we prepared Z-stack surface rendered 3-D models for P. aeruginosa (Fig. 7) and S. aureus at days 7, 14 and 21 (Fig. 8). Bacterial viability was also quantified at the three time points (day 7, 14 and 21) using 3D rendered biovolume Z-stack images obtained from the same samples used for single-plane fluorescence imaging. Each of the three time points represented an additional week of biofilm colonisation for P. aeruginosa (Fig. 7H) and S. aureus (Fig. 8H). P. aeruginosa incubated on the CTL samples showed significantly greater biovolume compared to the HTE samples at days 7, 14 and 21 (p = 0.003, p = 0.006 and p = 0.003 respectively, Fig. 7G). There was minimal evidence of clusters of live bacteria, associated with the formation of biofilm [64], on the HTE surfaces. Rather, there were thick areas of dead bacteria evident on these samples, most prominent at days 14 and 21. Although there was some increase in P. aeruginosa biovolume on the HTE surface by day 21, this was still less than half the biovolume on the CTL surface at day 7. While the viability of P. aeruginosa remained above 90% on the CTL surfaces out to day 21, from day 7 to day 14, the viability on the HTE surface remained below 25% (21.1 ± 3.7% at day 7, decreasing to 18.1 ± 3.5% at day 14), with most cells in contact with the surface killed at day 21. Interestingly, there was evidence of minor bacterial recolonisation above the previously killed bacteria on the HTE surface, which may partially explain the slight recovery in P. aeruginosa viability through the biovolume to 42.4 ± 5.2% by day 21 (Fig. 7H). Similarly, to P. aeruginosa, there was significantly less S. aureus biovolume on the HTE surfaces compared to the CTL surfaces at days 7, 14 and 21 (p = 0.002, p = 0.01 and p = 0.01 respectively, Fig. 8G). Again, by day 21, there was less than half the biovolume of S. aureus on the HTE surface compared to that on the CTL surface at day 7. Pockets of S. aureus recolonisation were evident away from the surface, particularly noticeable on days 14 and 21 (Fig. 8E and F), where total viability increased from 35.0 ± 6.2% at day 7, 53.1 ± 3.0% at day 14, to 63.4 ± 4.2% at day 21 (Fig. 8H).
Fig. 7.
Representative Z-stack rendered biofilm 3D-images of P. aeruginosa on CTL and HTE surfaces for day 7 (Fig. 7A and D, respectively), 14 (Fig. 7B and E, respectively) and 21 (Fig. 7C and F, respectively). Biovolume of P. aeruginosa incubated on CTL and HTE for day 7, 14 and 21 (Fig. 7G) mean ± SD, (n = 3). Yellow arrows highlight pockets of bacterial recolonisation on top of dead biofilm (scale bar = 30 μm), images signify an area of 0.01 mm2 (n = 3). Viability of P. aeruginosa (Fig. 7H) at three time points calculated from biovolume using multiple t-tests followed by the Holm-Sidak method, with alpha = 0.05. Each column represents mean ± SD, (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 8.
Representative Z-stack rendered biofilm 3D-images of S. aureus on CTL and HTE samples for day 7 (Fig. 8A and D, respectively), 14 (Fig. 8B and E, respectively) and 21 (8C and 8F, respectively). Scale bar = 30 μm. Biovolume of S. aureus incubated on CTL and HTE for day 7, 14 and 21 (6G) Mean ± SD, n = 3. Yellow arrows depict pockets of bacterial recolonisation on top of dead bacteria (scale bar 30 μm), images signify an area of 0.01 mm2 (n = 3). Viability of S. aureus (8H) at three time points calculated from biovolume using multiple t-tests followed by the Holm-Sidak method, with alpha = 0.05. Each column represents mean ± SD, (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
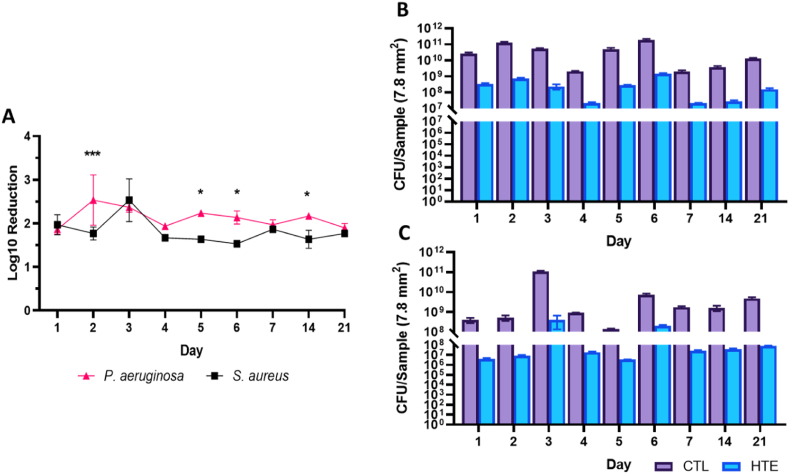
Using the drop plate method, the HTE surface showed a 1.9–2.5 log reduction in CFUs for P. aeruginosa, with the highest log reduction recorded on day 2 (2.5 log reduction). In contrast, S. aureus displayed a 1.5–2.5 log reduction in CFUs (Fig. 9A). The maximum log reduction for S. aureus on the HTE surface was on day 3, consistent with data from live/dead staining (Fig. 6B).
Fig. 9.
Log reduction of P. aeruginosa and S. aureus incubated on HTE surface over 21 days. Mean ± SD, (n = 3) and ∗ = p < 0.05 and ∗∗∗ = p < 0.001 (9A). CFUs per sample for P. aeruginosa (9B) and S. aureus (9C) on CTL and HTE surface mean ± SD, (n = 3). Table of p-values can be found for Fig. 9A, B and 9C in the supplementary information (Table S5, Table S6 and Table S7, respectively).
After day 3, the HTE samples incubated with S. aureus showed a consistently lower log reduction compared to P. aeruginosa until the end of the study, particularly on days 5, 6 and 14 (p < 0.05). Both P. aeruginosa and S. aureus showed CFU counts in the range of 2–4 orders of magnitude higher on the CTL than on the HTE surface throughout the 21-day study (Fig. 9B and C). There was a notable increase in log reduction on S. aureus incubated on HTE samples at day 3, in agreement with live/dead viability (Fig. 5, Fig. 6B). The unexpected finding that S. aureus showed a delayed maximum killing on the HTE surface at day 3 could be a function of the difference in membrane structure between Gram-positive and Gram-negative bacteria [65].
3.5. Morphology of P. aeruginosa and S. aureus on CTL and HTE surfaces
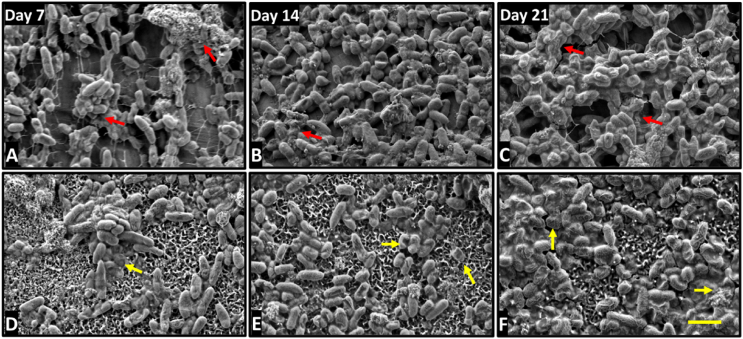
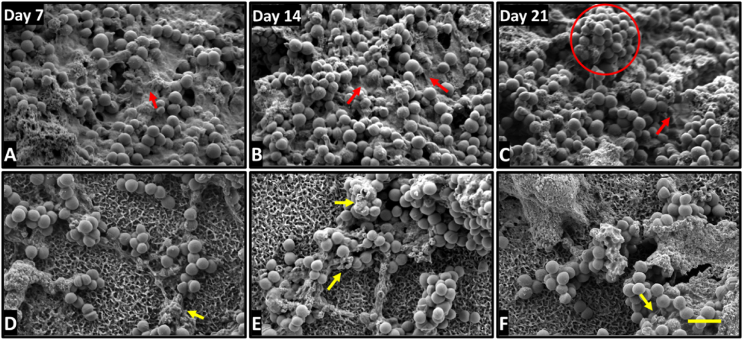
To gain further insights into differences between the interaction of P. aeruginosa and S. aureus with CTL and HTE surfaces over time, SEM micrographs were acquired over 21 days. For bacteria incubated on the HTE surface, there were distinct differences in morphology from days 7–21 compared to CTL surfaces. P. aeruginosa and S. aureus had almost completely colonised the CTL surfaces by day 7, with increasing cell density and little evidence of cell damage by day 21. The morphology of P. aeruginosa remained non-deformed, regular rod-shaped, attached to the surface by pili on CTL surfaces (Fig. 10, A-C). Similarly, S. aureus incubated on CTL surfaces, appeared spherical, with little evidence of membrane disruption, increasing in density and surface coverage, with notable mushroom-shaped formations indicative of biofilm formation by day 21 (Fig. 11, A-C) [64]. Further evidence that a mature biofilm had formed on the CTL surfaces by day 7 is the extensive extra polymeric substance (EPS) [66], which increased in density by day 21. In contrast, both bacterial species remained in small clusters on the HTE surfaces out to day 21, with continued evidence of cell rupture. Examples of lysed, flattened, and fragmented P. aeruginosa can be seen in Fig. 10, D-E, and the ruffled and fragmented morphology of S. aureus can be seen in Fig. 11, D-E. The inset images in Figure S1 demonstrate membrane piercing by the nanostructures, consistent with other reports where both Gram-negative and Gram-positive bacteria have been killed on comparable surfaces at earlier timepoints [37,67]. As confirmed by live/dead viability assays (Fig. 4, Fig. 6A) and CFU counts (Fig. 9), P. aeruginosa appeared to be inflicted with more cellular damage than S. aureus due to the nanostructures on the HTE surface. Significantly, we observed minimal EPS produced by both bacteria and fewer cells on the HTE surfaces compared to CTL samples at all timepoints (Figs. 10 and 11, A-C). During biofilm formation EPS is vital to the shift in cell state from adhered planktonic cells to a recalcitrant mature biofilm, where bacteria within the biofilm are more resistant to antibacterial drugs and host immune responses [68,69]. Cao et al. [70] reported similar findings on nanostructured titanium alloy towards S. epidermidis, where biofilm formation was delayed for up to 6 days. However, to the authors’ knowledge this is the first study that demonstrates delayed biofilm formation for P. aeruginosa and S. aureus on a nanostructured surface for 21 days.
Fig. 10.
SEM micrographs of P. aeruginosa incubated for 7, 14 and 21 days on CTL (A–C) and HTE surfaces (D–F). Yellow arrows are highlighting irregular morphology and dead cells: Red arrows show EPS covering bacteria. Scale bar bottom left = 2 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 11.
SEM micrographs of S. aureus incubated for 7, 14 and 21 days on CTL (A–C) and HTE surfaces (D–F). Arrows are highlighting irregular morphology and dead cells; Red arrows show EPS covering bacteria: Red circle highlights the formation of mushroom-shaped biofilm. Scale bar bottom left = 2 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Bioinspired nanostructured surfaces have previously been reported to show antibacterial efficacy against pathogens involved in IAIs [34,35,38,71]. However, this is the first study to examine the efficacy of such a surface for extended periods of time. We evaluated the efficacy of the nanostructures against two pathogens commonly associated with IAIs, i.e., the Gram-negative P. aeruginosa and the Gram-positive S. aureus, over the period of 21 days. We observed a sustained high killing rate on the HTE surface for both P. aeruginosa and S. aureus of approximately 97% and 60% by live/dead analysis at day 21, respectively. Analysis of biovolume demonstrated that P. aeruginosa and S. aureus remained only approximately 60% and 40% viable by day 21, respectively, where most live bacteria were away from the surface. Conversely, P. aeruginosa and S. aureus incubated on CTL surfaces remained greater than 95% viable throughout the study, both at the interface with the surface and throughout the full thickness of the biomass. There was a consistent 2-log reduction in viability for both bacteria on the HTE surface compared to CTL throughout the study. SEM images provided further evidence of a mechano-bactericidal effect of the spike-like nanostructures, also shown by other researchers at early timepoints [37,67], however our study showed that this effect was prolonged, with far fewer bacteria colonising the surface on the HTE samples compared to CTL samples out to day 21. By day 21, there remained a lack of EPS on HTE surfaces, where flattened, disrupted, and fragmented bacteria were observed. In contrast, both bacteria appeared to have formed mature biofilms on the CTL surfaces. Taken together, these results confirm our hypothesis that the antibacterial nanostructured implant surface disrupted biofilm growth and maturation over a long period.
Although the HTE surface was effective at killing both bacteria in this study, it appeared more so against P. aeruginosa, which may reflect the differences between Gram-negative and Gram-positive bacteria. It is well known that Gram-negative bacteria are less rigid than their Gram-positive counterparts, predominately due to the thinner peptidoglycan layer present in the cell wall, thus rendering them vulnerable to contact killing by the spike-like nanostructures [65,72]. Although Gram-positive bacteria lack an outer membrane, they are characterised by a thick peptidoglycan layer and protective capsular polysaccharide outer layer, providing greater cell wall rigidity and increased resistance to mechanically induced killing than Gram-negative bacteria [73,74].
The rate of cell division may also be a vital contributing factor to the difference in killing rate observed between Gram-negative and Gram-positive bacteria on the HTE surface. During cytokinesis, a complex hierarchical assembly of proteins form at the mid-cell known as the divisome [75]. This remodelling may alter the bacterial turgor pressure [76,77] and cell wall integrity [78,79], resulting in heightened killing on the HTE surface of bacteria which divide more rapidly. The doubling time for P. aeruginosa can be as low as 25 min [80], in contrast to 1 h for S. aureus [81]. The differences in doubling time between the two bacteria may be an additional influence on the killing rate on the HTE surface, reported by in this study and by other researchers [36,42,44,82].
Importantly, we observed substantial volumes of dead bacteria throughout the biomass, away from the HTE surface over 21 days. It is likely that not all these dead bacteria visible throughout the biomass came directly into contact with the nanostructured surface. Stress-induced programmed cell death (PCD) may provide an explanation for how a passive non-eluting surface may induce bacterial cell death at some distance away from the surface. PCD refers to the genetically encoded cascade of events leading to cell death, and may be initiated by cellular stress, for example oxidative stress [83,84]. Although PCD is a well understood phenomenon within eukaryotic cells, there are emerging studies that show PCD has been conserved throughout evolution in prokaryotic cells [85,86]. A biofilm may induce PCD within cells that are undergoing excessive environmental stresses, such as contact killing with a nanostructured surface, to favour the survival of the colony [86,87]. While the majority of the biovolume appeared to be constituted from dead bacteria on the HTE surfaces, there were observable pockets of live cells remerging on top of previously killed cells, particularly evident for S. aureus at day 14 and 21. Additionally, stressed cells in contact with the HTE surface may express extracellular death factor (EDF), which has been implicated in PCD in Gram-negative bacteria, mainly studied in E. coli [88,89].
Furthermore, gene expression may be modulated by the nanostructures, perhaps resulting in biofilm disaggregation. Quorum sensing genes, which synchronise the biofilm allowing it to behave like a multicellular organism, may be modulated, thus affecting biofilm formation over time. Potential down regulation in quorum sensing and biofilm associated genes may inhibit formation of biofilm on the HTE surface, though this warrants further investigation. It is obvious that unravelling a complete mechanistic picture of the effect of surface nanostructures on bacterial attachment, growth, colonisation, and biofilm establishment is a complex task that will require the efforts of much ongoing research. However, this work details pioneering experiments which demonstrate the capacity of surfaces containing disordered spike-like nanostructures to sustain antibacterial properties over an extended period. Clinically, such an implant surface may not only act as a deterrent to bacterial aggregation and biofilm formation, but it may also provide host tissue with more time to win the race to the surface, in turn reducing the likelihood of an IAI.
4. Conclusion
In this study, we characterised the physico-chemical and antibacterial properties of hydrothermally etched nanostructures over extended time periods. A short-term cytocompatibility study using RAW 264.7 macrophage-like cells showed no adverse effects on morphology, viability, and proliferation. Following culture on HTE surfaces, P. aeruginosa did not exceed 4% viability over a 21-day period, whereas S. aureus viability reduced to 6.8 ± 3.1% by day 3, then increased to approximately 40% by day 21. In contrast, both bacterial species remained above 95% viable on CTL surfaces over the duration of the study. Total biovolume and measured viability was also significantly lower on the HTE surfaces for both bacteria at all timepoints. SEM imaging confirmed contact killing of bacteria on the nanostructures, as well as a lack of mature, organised biofilm forming on the HTE surfaces over longer timepoints. Although additional research is required to fully understand the mechanisms underpinning the long-term antibacterial properties of surfaces containing sharp nanostructures, including an in vivo model, this study points to the potential of such surfaces to be applied to medical devices and ultimately contribute to reducing the rates of IAI.
Author contributions
R.B., and D.F. designed, planned, and performed the experiments. R.B. analysed data and drafted the manuscript. D.F. fabricated samples. J.W., A.B., N.N. and D.P. performed the surface characterisation and participated in data analysis, D.F., D.B. and T.B. initiated the design of the study and revised the manuscript. K.V. supervised the study and revised the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author, [KV], upon reasonable request.
Declaration of competing interest
The authors listed above declare no conflict of interest or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was co-funded by the Department of Industry, Science, Energy and Resources (Innovative Manufacturing CRC Ltd) Global Orthopaedic Technology Pty Ltd (IMCRC/GOT/130318). The authors acknowledge the funding and in-kind support from Corin Australia and the University of South Australia. The authors would also like to acknowledge the instruments and scientific and technical assistance of Microscopy Australia at the University of South Australia, Mawson Lakes Campus, a facility that is funded by the University, and State and Federal Governments. RB thanks Andrew Hayles for his assistance in the graphical abstract. KV thanks NHMRC for Fellowship GNT1194466 and ARC. for grant DP180101254.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100176.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Additional information regarding the following: Characterisation by SEM, AFM, and wettability (Tables S1), XPS elemental analysis mean and p-values for CTL and HTE surfaces (Table S2). A table of p-values for live/dead viability analysis for P. aeruginosa (Table S3) and S. aureus (Table S4) from Fig. 5A and B respectively. Mean log-reduction scores and associated p-values (Table S5) from Fig. 9A. P-values for percent viability calculated from colony forming unit analysis for P. aeruginosa (Table S6) and S. aureus (Table S7), from Fig. 9B and C respectively. Lastly, Figure S1 SEM micrographs for P. aeruginosa from Fig. 10 and S. aureus from Fig. 11, incubated on HTE surface at day 7. Inset images depict membrane piercing and deformed morphology.
References
- 1.Long P.H. Medical devices in orthopedic applications. Toxicol. Pathol. 2008;36(1):85–91. doi: 10.1177/0192623307310951. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Q., Mao L., Shi Y., Fu W., Hu Y. Biocompatibility of Ti-6Al-4V titanium alloy implants with laser microgrooved surfaces. Mater. Technol. 2020:1–10. [Google Scholar]
- 3.Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350(14):1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 4.Moriarty T.F., Kuehl R., Coenye T., Metsemakers W.J., Morgenstern M., Schwarz E.M., Riool M., Zaat S.A.J., Khana N., Kates S.L., Richards R.G. Orthopaedic device-related infection: current and future interventions for improved prevention and treatment. EFORT Open Rev. 2016;1(4):89–99. doi: 10.1302/2058-5241.1.000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patzakis M.J., Wilkins J. Factors influencing infection rate in open fracture wounds. Clin. Orthop. Relat. Res. 1989;243:36–40. [PubMed] [Google Scholar]
- 6.Boxma H., Broekhuizen T., Patka P., Oosting H. Randomised controlled trial of single-dose antibiotic prophylaxis in surgical treatment of closed fractures: the Dutch Trauma Trial. Lancet. 1996;347(9009):1133–1137. doi: 10.1016/s0140-6736(96)90606-6. [DOI] [PubMed] [Google Scholar]
- 7.Hernández-Vaquero D., Fernández-Fairen M., Torres A., Menzie A.M., Fernández-Carreira J.M., Murcia-Mazon A., Guerado E., Merzthal L. Treatment of periprosthetic infections: an economic analysis. ScientificWorldJournal. 2013;2013:821650. doi: 10.1155/2013/821650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haddad F.S., Ngu A., Negus J.J. Prosthetic joint infections and cost analysis? Adv. Exp. Med. Biol. 2017;971:93–100. doi: 10.1007/5584_2016_155. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J.M. Future challenges in the in vitro and in vivo evaluation of biomaterial biocompatibility. Regen Biomater. 2016;3(2):73–77. doi: 10.1093/rb/rbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gristina A.G. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science. 1987;237(4822):1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- 11.Chu L., Yang Y., Yang S., Fan Q., Yu Z., Hu X.L., James T.D., He X.P., Tang T. Preferential colonization of osteoblasts over Co-cultured bacteria on a bifunctional biomaterial surface. Front. Microbiol. 2018;9:2219. doi: 10.3389/fmicb.2018.02219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roilides E., Simitsopoulou M., Katragkou A., Walsh T.J. How biofilms evade host defenses. Microbiol. Spectr. 2015;3(3) doi: 10.1128/microbiolspec.MB-0012-2014. [DOI] [PubMed] [Google Scholar]
- 13.Hall C.W., Mah T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017;41(3):276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 14.Mah T.F., O'Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 15.Seebach E., Kubatzky K.F. Chronic implant-related bone infections-can immune modulation be a therapeutic strategy? Front. Immunol. 2019;10:1724. doi: 10.3389/fimmu.2019.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trebse R., Pisot V., Trampuz A. Treatment of infected retained implants. J Bone Joint Surg Br. 2005;87(2):249–256. doi: 10.1302/0301-620x.87b2.15618. [DOI] [PubMed] [Google Scholar]
- 17.Schömig F., Perka C., Pumberger M., Ascherl R. Implant contamination as a cause of surgical site infection in spinal surgery: are single-use implants a reasonable solution? - a systematic review. BMC Muscoskel. Disord. 2020;21(1):634. doi: 10.1186/s12891-020-03653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue C., Zhao B., Ren Y., Kuijer R., van der Mei H.C., Busscher H.J., Rochford E.T. The implant infection paradox: why do some succeed when others fail? Opinion and discussion paper. Eur. Cell. Mater. 2015;29:303–310. doi: 10.22203/ecm.v029a23. ; discussion 310-3. [DOI] [PubMed] [Google Scholar]
- 19.Schierholz J.M., Beuth J. Implant infections: a haven for opportunistic bacteria. J. Hosp. Infect. 2001;49(2):87–93. doi: 10.1053/jhin.2001.1052. [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro M., Monteiro F.J., Ferraz M.P. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter. 2012;2(4):176–194. doi: 10.4161/biom.22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brouqui P., Rousseau M.C., Stein A., Drancourt M., Raoult D. Treatment of Pseudomonas aeruginosa-infected orthopedic prostheses with ceftazidime-ciprofloxacin antibiotic combination. Antimicrob. Agents Chemother. 1995;39(11):2423–2425. doi: 10.1128/aac.39.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crémet L., Broquet A., Brulin B., Jacqueline C., Dauvergne S., Brion R., Asehnoune K., Corvec S., Heymann D., Caroff N. Pathogenic potential of Escherichia coli clinical strains from orthopedic implant infections towards human osteoblastic cells. Pathog Dis. 2015;73(8):ftv065. doi: 10.1093/femspd/ftv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cogen A.L., Nizet V., Gallo R.L. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 2008;158(3):442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobolev A., Valkov A., Kossenko A., Wolicki I., Zinigrad M., Borodianskiy K. Bioactive coating on Ti alloy with high osseointegration and antibacterial Ag nanoparticles. ACS Appl. Mater. Interfaces. 2019;11(43):39534–39544. doi: 10.1021/acsami.9b13849. [DOI] [PubMed] [Google Scholar]
- 25.Haidari H., Kopecki Z., Bright R., Cowin A.J., Garg S., Goswami N., Vasilev K. Ultrasmall AgNP-impregnated biocompatible hydrogel with highly effective biofilm elimination properties. ACS Appl. Mater. Interfaces. 2020;12(37):41011–41025. doi: 10.1021/acsami.0c09414. [DOI] [PubMed] [Google Scholar]
- 26.RavindranGirija A., Balasubramanian S., Bright R., Cowin A.J., Goswami N., Vasilev K. Ultrasmall gold nanocluster based antibacterial nanoaggregates for infectious wound healing. ChemNanoMat. 2019;5(9):1176–1181. [Google Scholar]
- 27.Riool M., de Breij A., Drijfhout J.W., Nibbering P.H., Zaat S.A.J. Antimicrobial peptides in biomedical device manufacturing. Front Chem. 2017;5:63. doi: 10.3389/fchem.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griesser S.S., Jasieniak M., Vasilev K., Griesser H.J. Antimicrobial peptides grafted onto a plasma polymer interlayer platform: performance upon extended bacterial challenge. Coatings. 2021;11(1):68. [Google Scholar]
- 29.Jahanmard F., Dijkmans F.M., Majed A., Vogely H.C., van der Wal B.C.H., Stapels D.A.C., Ahmadi S.M., Vermonden T., Amin Yavari S. Toward antibacterial coatings for personalized implants. ACS Biomater. Sci. Eng. 2020;6(10):5486–5492. doi: 10.1021/acsbiomaterials.0c00683. [DOI] [PubMed] [Google Scholar]
- 30.Vasilev K. Nanoengineered antibacterial coatings and materials: a perspective. Coatings. 2019;9(10):654. [Google Scholar]
- 31.Vasilev K., Cook J., Griesser H.J. Antibacterial surfaces for biomedical devices. Expet Rev. Med. Dev. 2009;6(5):553–567. doi: 10.1586/erd.09.36. [DOI] [PubMed] [Google Scholar]
- 32.Suhardi V.J., Bichara D.A., Kwok S., Freiberg A.A., Rubash H., Malchau H., Yun S.H., Muratoglu O.K., Oral E. A fully functional drug-eluting joint implant. Nat Biomed Eng. 2017;1 doi: 10.1038/s41551-017-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eltorai A.E., Haglin J., Perera S., Brea B.A., Ruttiman R., Garcia D.R., Born C.T., Daniels A.H. Antimicrobial technology in orthopedic and spinal implants. World J. Orthoped. 2016;7(6):361–369. doi: 10.5312/wjo.v7.i6.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanova E.P., Hasan J., Webb H.K., Gervinskas G., Juodkazis S., Truong V.K., Wu A.H., Lamb R.N., Baulin V.A., Watson G.S., Watson J.A., Mainwaring D.E., Crawford R.J. Bactericidal activity of black silicon. Nat. Commun. 2013;4:2838. doi: 10.1038/ncomms3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bright R., Hayles A., Fernandes D., Visalakshan R.M., Ninan N., Palms D., Burzava A., Barker D., Brown T., Vasilev K. In vitro bactericidal efficacy of nanostructured Ti6Al4V surfaces is bacterial load dependent. ACS Appl. Mater. Interfaces. 2021;13(32):38007–38017. doi: 10.1021/acsami.1c06919. [DOI] [PubMed] [Google Scholar]
- 36.Wandiyanto J.V., Tamanna T., Linklater D.P., Truong V.K., Al Kobaisi M., Baulin V.A., Joudkazis S., Thissen H., Crawford R.J., Ivanova E.P. Tunable morphological changes of asymmetric titanium nanosheets with bactericidal properties. J. Colloid Interface Sci. 2020;560:572–580. doi: 10.1016/j.jcis.2019.10.067. [DOI] [PubMed] [Google Scholar]
- 37.Ivanova E.P., Linklater D.P., Werner M., Baulin V.A., Xu X., Vrancken N., Rubanov S., Hanssen E., Wandiyanto J., Truong V.K., Elbourne A., Maclaughlin S., Juodkazis S., Crawford R.J. The multi-faceted mechano-bactericidal mechanism of nanostructured surfaces. Proc. Natl. Acad. Sci. U. S. A. 2020;117(23):12598–12605. doi: 10.1073/pnas.1916680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linklater D.P., De Volder M., Baulin V.A., Werner M., Jessl S., Golozar M., Maggini L., Rubanov S., Hanssen E., Juodkazis S., Ivanova E.P. High aspect ratio nanostructures kill bacteria via storage and release of mechanical energy. ACS Nano. 2018;12(7):6657–6667. doi: 10.1021/acsnano.8b01665. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins J., Mantell J., Neal C., Gholinia A., Verkade P., Nobbs A.H., Su B. Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 2020;11(1):1626. doi: 10.1038/s41467-020-15471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eaninwene G., 2nd, Yao C., Webster T.J. Enhanced osteoblast adhesion to drug-coated anodized nanotubular titanium surfaces. Int. J. Nanomed. 2008;3(2):257–264. doi: 10.2147/ijn.s2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nichol T., Callaghan J., Townsend R., Stockley I., Hatton P.V., Le Maitre C., Smith T.J., Akid R. The antimicrobial activity and biocompatibility of a controlled gentamicin-releasing single-layer sol-gel coating on hydroxyapatite-coated titanium. Bone Joint Lett. J. 2021;103-b(3):522–529. doi: 10.1302/0301-620X.103B3.BJJ-2020-0347.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vishnu J., V K.M., Gopal V., Bartomeu Garcia C., Hameed P., Manivasagam G., Webster T.J. Hydrothermal treatment of etched titanium: a potential surface nano-modification technique for enhanced biocompatibility. Nanomedicine. 2019;20:102016. doi: 10.1016/j.nano.2019.102016. [DOI] [PubMed] [Google Scholar]
- 43.Ma K., Zhang R., Sun J., Liu C. Oxidation mechanism of biomedical titanium alloy surface and experiment. International Journal of Corrosion. 2020;2020:1678615. [Google Scholar]
- 44.Bhadra C.M., Truong V.K., Pham V.T., Al Kobaisi M., Seniutinas G., Wang J.Y., Juodkazis S., Crawford R.J., Ivanova E.P. Antibacterial titanium nano-patterned arrays inspired by dragonfly wings. Sci. Rep. 2015;5:16817. doi: 10.1038/srep16817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elliott D.T., Wiggins R.J., Dua R. Bioinspired antibacterial surface for orthopedic and dental implants. J. Biomed. Mater. Res. B Appl. Biomater. 2021;109(7):973–981. doi: 10.1002/jbm.b.34762. [DOI] [PubMed] [Google Scholar]
- 46.Ferraris S., Cochis A., Cazzola M., Tortello M., Scalia A., Spriano S., Rimondini L. Cytocompatible and anti-bacterial adhesion nanotextured titanium oxide layer on titanium surfaces for dental and orthopedic implants. Front Bioeng Biotechnol. 2019;7:103. doi: 10.3389/fbioe.2019.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiels S.M., Bedigrew K.M., Wenke J.C. Development of a hematogenous implant-related infection in a rat model. BMC Muscoskel. Disord. 2015;16:255. doi: 10.1186/s12891-015-0699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arciola C.R., Campoccia D., Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018;16(7):397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 49.Shi Q., Roux S., Latourte F., Hild F., Loisnard D., Brynaert N. Measuring topographies from conventional SEM acquisitions. Ultramicroscopy. 2018;191:18–33. doi: 10.1016/j.ultramic.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Kaja S., Payne A.J., Naumchuk Y., Koulen P. Quantification of lactate dehydrogenase for cell viability testing using cell lines and primary cultured astrocytes. Curr Protoc Toxicol. 2017;72:2.26.1–2.26.10. doi: 10.1002/cptx.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho M.H., Niles A., Huang R., Inglese J., Austin C.P., Riss T., Xia M. A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol. Vitro. 2008;22(4):1099–1106. doi: 10.1016/j.tiv.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S.S., Ghosh A.R. Assessment of bacterial viability: a comprehensive review on recent advances and challenges. Microbiology (Read.) 2019;165(6):593–610. doi: 10.1099/mic.0.000786. [DOI] [PubMed] [Google Scholar]
- 53.Bankier C., Cheong Y., Mahalingam S., Edirisinghe M., Ren G., Cloutman-Green E., Ciric L. A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLoS One. 2018;13(2) doi: 10.1371/journal.pone.0192093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canet-Ferrer J., Coronado E., Forment-Aliaga A., Pinilla-Cienfuegos E. Correction of the tip convolution effects in the imaging of nanostructures studied through scanning force microscopy. Nanotechnology. 2014;25(39):395703. doi: 10.1088/0957-4484/25/39/395703. [DOI] [PubMed] [Google Scholar]
- 55.Wenzel R.N. Surface roughness and contact angle. J. Phys. Colloid Chem. 1949;53(9):1466–1467. [Google Scholar]
- 56.Cassie A.B.D., Baxter S. Wettability of porous surfaces. Trans. Faraday Soc. 1944;40:546–551. 0. [Google Scholar]
- 57.Webb H.K., Crawford R.J., Ivanova E.P. Wettability of natural superhydrophobic surfaces. Adv. Colloid Interface Sci. 2014;210:58–64. doi: 10.1016/j.cis.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 58.Arango-Santander S., Pelaez-Vargas A., Freitas S.C., García C. A novel approach to create an antibacterial surface using titanium dioxide and a combination of dip-pen nanolithography and soft lithography. Sci. Rep. 2018;8(1):15818. doi: 10.1038/s41598-018-34198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabe M., Verdes D., Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv Colloid Interfac. 2011;162(1–2):87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Abaricia J.O., Shah A.H., Musselman R.M., Olivares-Navarrete R. Hydrophilic titanium surfaces reduce neutrophil inflammatory response and NETosis. Biomater Sci. 2020;8(8):2289–2299. doi: 10.1039/c9bm01474h. [DOI] [PubMed] [Google Scholar]
- 61.Sartoretto S.C., Calasans-Maia J.A., Costa Y.O.D., Louro R.S., Granjeiro J.M., Calasans-Maia M.D. Accelerated healing period with hydrophilic implant placed in sheep tibia. Braz. Dent. J. 2017;28(5):559–565. doi: 10.1590/0103-6440201601559. [DOI] [PubMed] [Google Scholar]
- 62.Kzhyshkowska J., Gudima A., Riabov V., Dollinger C., Lavalle P., Vrana N.E. Macrophage responses to implants: prospects for personalized medicine. J. Leukoc. Biol. 2015;98(6):953–962. doi: 10.1189/jlb.5VMR0415-166R. [DOI] [PubMed] [Google Scholar]
- 63.Stiefel P., Schmidt-Emrich S., Maniura-Weber K., Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15:36. doi: 10.1186/s12866-015-0376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bjarnsholt T., Alhede M., Alhede M., Eickhardt-Sørensen S.R., Moser C., Kühl M., Jensen P., Høiby N. The in vivo biofilm. Trends Microbiol. 2013;21(9):466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 65.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5) doi: 10.1101/cshperspect.a000414. a000414-a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., Xiao P., Wang Y., Hao Y. Mechanisms and control measures of mature biofilm resistance to antimicrobial agents in the clinical context. ACS Omega. 2020;5(36):22684–22690. doi: 10.1021/acsomega.0c02294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linklater D.P., Baulin V.A., Juodkazis S., Crawford R.J., Stoodley P., Ivanova E.P. Mechano-bactericidal actions of nanostructured surfaces. Nat. Rev. Microbiol. 2021;19(1):8–22. doi: 10.1038/s41579-020-0414-z. [DOI] [PubMed] [Google Scholar]
- 68.Aggarwal S., Stewart P.S., Hozalski R.M. Biofilm cohesive strength as a basis for biofilm recalcitrance: are bacterial biofilms overdesigned? Microbiol. Insights. 2015;8(Suppl 2):29–32. doi: 10.4137/MBI.S31444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.González J.F., Hahn M.M., Gunn J.S. Chronic biofilm-based infections: skewing of the immune response. Pathogens and disease. 2018;76(3):fty023. doi: 10.1093/femspd/fty023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao Y., Su B., Chinnaraj S., Jana S., Bowen L., Charlton S., Duan P., Jakubovics N.S., Chen J. Nanostructured titanium surfaces exhibit recalcitrance towards Staphylococcus epidermidis biofilm formation. Sci. Rep. 2018;8(1):1071. doi: 10.1038/s41598-018-19484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripathy A., Sen P., Su B., Briscoe W.H. Natural and bioinspired nanostructured bactericidal surfaces. Adv. Colloid Interface Sci. 2017;248:85–104. doi: 10.1016/j.cis.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnoldi M., Fritz M., Bäuerlein E., Radmacher M., Sackmann E., Boulbitch A. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E. 2000;62(1 Pt B):1034–1044. doi: 10.1103/physreve.62.1034. [DOI] [PubMed] [Google Scholar]
- 73.O'Riordan K., Lee J.C. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 2004;17(1):218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harper C.E., Hernandez C.J. Cell biomechanics and mechanobiology in bacteria: challenges and opportunities. APL Bioeng. 2020;4(2) doi: 10.1063/1.5135585. 021501-021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bohuszewicz O., Liu J., Low H.H. Membrane remodelling in bacteria. J. Struct. Biol. 2016;196(1):3–14. doi: 10.1016/j.jsb.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osawa M., Erickson H.P. Turgor pressure and possible constriction mechanisms in bacterial division. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00111. 111-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coltharp C., Buss J., Plumer T.M., Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci. U. S. A. 2016;113(8):E1044–E1053. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egan A.J., Cleverley R.M., Peters K., Lewis R.J., Vollmer W. Regulation of bacterial cell wall growth. FEBS J. 2017;284(6):851–867. doi: 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- 79.Strahl H., Hamoen L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 2010;107(27):12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaBauve A.E., Wargo M.J. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter. 2012;6 doi: 10.1002/9780471729259.mc06e01s25. Unit-6E.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szafrańska A.K., Junker V., Steglich M., Nübel U. Rapid cell division of Staphylococcus aureus during colonization of the human nose. BMC Genom. 2019;20(1):229. doi: 10.1186/s12864-019-5604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsimbouri P.M., Fisher L., Holloway N., Sjostrom T., Nobbs A.H., Meek R.M., Su B., Dalby M.J. Osteogenic and bactericidal surfaces from hydrothermal titania nanowires on titanium substrates. Sci. Rep. 2016;6:36857. doi: 10.1038/srep36857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong Y., Zeng J., Wang X., Drlica K., Zhao X. Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 2019;116(20):10064–10071. doi: 10.1073/pnas.1901730116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ezraty B., Gennaris A., Barras F., Collet J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017;15(7):385–396. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 85.Bayles K.W. Bacterial programmed cell death: making sense of a paradox. Nat. Rev. Microbiol. 2014;12(1):63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allocati N., Masulli M., Di Ilio C., De Laurenzi V. Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis. 2015;6(1) doi: 10.1038/cddis.2014.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Engelberg-Kulka H., Sat B., Reches M., Amitai S., Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12(2):66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 88.Belitsky M., Avshalom H., Erental A., Yelin I., Kumar S., London N., Sperber M., Schueler-Furman O., Engelberg-Kulka H. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. Cell. 2011;41(6):625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 89.Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2(10) doi: 10.1371/journal.pgen.0020135. e135-e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [KV], upon reasonable request.