Abstract
Objective
To analyze the genome-wide epigenomic and transcriptomic changes induced by long term resistance or endurance training in the hippocampus of wild-type mice.
Methods
We performed whole-genome bisulfite sequencing (WGBS) and RNA sequencing (RNA-seq) of mice hippocampus after 4 weeks of specific training. In addition, we used a novel object recognition test before and after the intervention to determine whether the exercise led to an improvement in cognitive function.
Results
Although the majority of DNA methylation changes identified in this study were training-model specific, most were associated with hypomethylation and were enriched in similar histone marks, chromatin states, and transcription factor biding sites. It is worth highlighting the significant association found between the loss of DNA methylation in Tet1 binding sites and gene expression changes, indicating the importance of these epigenomic changes in transcriptional regulation. However, endurance and resistance training activate different gene pathways, those being associated with neuroplasticity in the case of endurance exercise, and interferon response pathways in the case of resistance exercise, which also appears to be associated with improved learning and memory functions.
Conclusions
Our results help both understand the molecular mechanisms by which different exercise models exert beneficial effects for brain health and provide new potential therapeutic targets for future research.
Keywords: Epigenome, Transcriptome, Exercise, Resistance training, Endurance training, Hippocampus, Neuroplasticity
Highlights
-
•
Long-term training rewires the epigenome and transcriptome of mouse hippocampus.
-
•
Loss of DNA methylation at Tet1 binding sites plays a key role in the regulation of gene expression after training.
-
•
Different gene expression programs are activated in the hippocampus following endurance or resistance training models.
1. Introduction
The beneficial effects of exercise on human health have been known for a long time [[1], [2], [3]]. Exercise is well established as a healthy strategy that contributes to the prevention and delay in the onset of cardiovascular and metabolic diseases, as well as some types of cancers [[4], [5], [6], [7], [8]]. Moreover, exercise has been proven as an effective tool for brain health, having a protective role in both psychiatric disorders and neurodegenerative diseases (reviewed in [9]). In this regard, the impact of exercise on the brain has been extensively studied in the hippocampus, a region involved in learning and memory. In fact, hippocampal volume is reduced by 1–2% annually in older adults without dementia, increasing their risk of developing cognitive impairment [10]. This atrophy precedes and leads to memory impairment in later stages of aging. The hippocampus is particularly vulnerable to the harmful effects of the most prevalent chronic diseases [11,12] that have been associated with increased risk of developing Alzheimer's disease [13], the most common form of dementia.
However, several studies have shown that endurance (aerobic) exercise increases hippocampal volume in elderly people following interventions of between 3 and 12 months, indicating that hippocampal size is modifiable and recoverable at these ages [10,14]. The less explored area of resistance exercise has also been demonstrated to have beneficial effects that delay cognitive decline (reviewed in [15]). To this end, both models of exercise, although presenting significant differences at the physiological, metabolic, and molecular level [16], have demonstrated an ability to maintain brain health. In fact, by means of mouse models, it is possible to analyze the underlying molecular mechanisms associated with exercise. For instance, adult neurogenesis in the dentate gyrus of the hippocampus is one of the most studied brain responses to exercise in murine models. Recently, we have shown that the induction of neurogenesis is independent of the exercise model (endurance or resistance) [17].
Much remains to be explored to better understand the molecular mechanisms by which exercise is beneficial. Epigenetics is indeed a good molecular link to explain the effects of the environment, including exercise, on genes [18]. Several studies have shown that there are epigenetic changes in DNA and post-translational histone marks associated with appropriate hippocampus functioning [[19], [20], [21], [22], [23], [24], [25]], and some epigenetic alterations have also even been identified in neurodegenerative disorders such as Alzheimer's disease [26,27].
In this work, we performed whole-genome bisulfite sequencing (WGBS) and RNA sequencing (RNA-seq) of the hippocampus of wild-type mice subjected to long-term resistance or endurance training. We further analyzed and integrated the epigenomic and transcriptomic data from both models of exercise to identify common and specific changes for each type of training, and to better understand the mechanisms by which exercise has beneficial effects on this structure, which is involved in learning and memory processes.
2. Methods
2.1. Animals and experimental design
A total of 63 C57BL/6N male mice (8 weeks old) were randomly divided into three different groups: sedentary control (CTL, n = 24), resistance training (RES, n = 20), and endurance training (END, n = 20) (Figure S1). Mice were maintained on a 12-h light/dark cycle (onset 8:00 AM) and under controlled temperature (22 ± 2 °C) at the Animal Facilities of the University of Oviedo, Spain (authorized facility No. ES330440003591). All procedures were conducted early in light portion of the cycle and performed in accordance with the institutional guidelines approved by The Research Ethics Committee of the University of Oviedo, Spain (PROAE 10/2016). Mice were fed a pellet rodent diet (Teklad Irradiated Global 18% Protein Rodent Diet, Envigo, Spain) and water ad libitum. Food intake and animal weight were measured weekly.
2.2. Training devices
Endurance training was performed on a four-lane commercial treadmill (TSE Systems, Germany), with adjustable speed and slope and without any aversive stimuli. Resistance training was carried out using an in-house-manufactured ladder [17] consisting of 25 steel wire steps of 1.5 mm of diameter separated by 15 mm. A resting area of 20 × 20 cm was placed on top of the ladder. The slope of the ladder was modifiable, ranging between 90° and 80° with the horizontal plane.
2.3. Training protocols
The same researcher handled and trained the animals during each of the stages of training: acclimation period, physical performance tests (pre- and post-training), and training protocols.
2.4. Acclimation period
All mice were acclimatized to the training devices for 3 weeks, training for 15 min per device per day, 5 days/week [28]. Training load was kept to a minimum, avoiding adaptations that could interfere with pre-training maximal performance tests [28].
During the first week, mice were placed on the stopped treadmill and on the resting area at the top of the ladder. In the second and third weeks, mice walked on the moving treadmill belt (10 cm/s) and they were trained how to climb the ladder, from the 5th top step to the resting area, the number of rungs climbed gradually increasing to 10.
The use of aversive stimuli to encourage treadmill running or ladder climbing were avoided for the whole period [[28], [29], [30], [31]]. To facilitate adaptation to endurance training, a static brush was used at the back end of the treadmill, to keep the animals running. In the case of resistance training, since different weights were to be attached to their tails with adhesive tape, a piece of tape was attached to their tails while climbing the ladder. After a few days, a light load (5 g) was attached to the mice's tails with tape. Mice were handled in groups of four, all of them sharing the same cage, to reduce anxiety.
This acclimation protocol allowed us to train all the animals without refusals [30].
2.5. Maximal performance tests
Forty-eight hours after the end of the acclimation period, mice were randomly distributed in the above-mentioned groups. Mice in the CTL group performed maximal tests for endurance and resistance capacities, while the mice in the RES and END groups only underwent the tests corresponding to the quality they were going to be trained for.
Maximal endurance capacity was determined by an incremental test on the treadmill, adapted from other studies [32,33]. In short, after a 10-min warm-up at 15 cm/s with 10° slope, the incremental test started at 20 cm/s. Every 3 min, speed was increased by 5 cm/s, until exhaustion. Maximum speed (cm/s) and total time (min) were recorded. Total time was used as a measurement of maximal endurance capacity.
Maximal resistance capacity was tested using the vertical ladder, following a protocol adapted from previous studies [34]. Mice performed a warm-up consisting of 3 series of 10 repetitions, 10 steps/repetition, at 90° slope, with no external load. The animals rested for 60 s between series. Then, the slope was set at 85° and the animals performed successive series of 10 steps with increasing external loads until exhaustion. The initial external load was 10 g, increasing by 5 g in each series. The mice rested for 120 s in the resting area after each series. If the animals failed to climb 10 steps with a certain weight load, they were allowed to try again with the same load after 120 s of rest. If they failed again, the weight load of the last complete series was recorded as their maximal weight load. The maximal resistance capacity was then expressed as the maximal weight load relative to body weight.
Both tests were repeated at the end of the training period, following the same protocols.
2.6. Training protocols
All END and RES mice trained for 4 weeks, 5 days/week (Monday to Friday). Training protocols were adapted from previous works [17,28] in terms of the intensity and duration of sessions.
Endurance training sessions started with an identical warm-up as for the maximal endurance performance test. All sessions of continuous running had a mean duration of 60 min and the distance covered every day was 1000 m, as a fixed exercise volume. The intensity in terms of maximal speed, number of stages, as well as the speed and duration of each stage, varied throughout the week according to the following structure: 2 days at high intensity (Tuesday and Friday), 2 days at moderate intensity (Monday and Thursday), and 1 day at low intensity (Wednesday). Speed ranged from 40 to 80% of the mean maximal speed at the pre-training test [35]. The duration of each stage varied inversely with speed, between 15 and 5 min [35]. The slope was fixed at 10°. Maximal intensity increased throughout the training period, although the weekly schedule was maintained, as were the duration and the distance covered in training sessions.
Resistance training sessions also started with the same warm-up as for the maximal resistance performance test. All sessions were designed to achieve the same exercise volume by means of a combination of number of steps climbed (or distance against gravity) and weight load [36]. Considering the combination of these parameters, accumulated work of 260 mJ (g·m2/s2) was achieved daily. The number of steps per training session varied between 400 and 2000 depending on the maximal weight load, which ranged between 25% and 65% of the maximal weight load at the pre-training test. We selected these maximum weight ranges because it has been reported that below 75% of 1 repetition maximum there is no velocity loss, which is important for standardizing the intensity of submaximal efforts [37]. The weekly schedule was: 2 days with high weight load and low number of steps (Tuesday and Friday), 2 days of intermediate weight load and number of steps (Monday and Thursday), and 1 day without weight load but a high number of steps (Wednesday). The number of steps and the maximum weight loads increased throughout the training period, whilst maintaining the weekly schedule, accumulated work, and percentages of maximal weight load.
Control mice remained in a cage in the same room as where END and RES animals were training.
2.7. Novel object recognition test
Mice performed a novel object recognition test (NOR) in the first week of the acclimation period (pre-training) and in the last week of the 4-week training period (post-training). NOR is a learning and memory test based on the natural disposition of mice to explore new environments [38]. The test was performed as previously described [17]. Briefly, mice (12 CTL, 12 RES, and 11 END) were placed in the center of a rectangular arena cage and left free to explore for 5 min as a habituation phase. Twenty-four hours later, a training phase was carried out whereby the mice were placed in the same area, where two identical objects were positioned opposite each other (objects A and A1) and given freedom of movement for 5 min. Ninety minutes later, object A1 was replaced by a new object, B, to determine the short-term memory (STM) capacity of the mice. Twenty-four hours after the STM phase, a second new object (C) replaced object B in order for long-term memory (LTM) to be analyzed. The arena was cleaned with 70% alcohol after each mouse. The whole process was recorded using a zenithal camera connected to a computer. Two researchers, blind as to the intervention group of the mouse involved, independently analyzed the videos from each of the phases and determined the number of times the mouse interacted with objects A, B and C. STM and LTM was calculated using the formula ((N-A)/(A + N) ∗ 100, where N is B for STM and C for LTM.
2.8. Tissue collection
Twenty-four hours after the final training session, mice were deeply anaesthetized with ketamine (80 mg/kg) and xylazine (5 mg/kg) in saline solution. Each mouse was perfused with phosphate buffer saline (PBS) via the abdominal vena cava and the brain was removed and sagittally dissected. The hippocampus was extracted from the right hemisphere, then rapidly frozen and stored at −80 °C until use. The left hemisphere was fixed with 4% paraformaldehyde in Sorensen's phosphate buffer overnight at 4 °C for histology processing.
2.9. Phospho-histone H3 immunostaining
Fixed left hemispheres were washed in PBS and immersed in 30% sucrose in PBS for 48 h for cryoprotection. Samples were then frozen in OCT (Optimal Cutting Temperature, Tissue-Tek) and stored at −80 °C until use. Sagittal sections (30 μm) were cut on an HM 350S microtome (Thermo Fisher Scientific In.) and stored in a solution containing 30% glycerol and 30% ethylene glycol in 0.02 M monobasic phosphate buffer of pH 7.2 at −20 °C.
For immunofluorescence, at least three non-consecutive sections for each mouse (3 CTL, 4 RES, and 4 END) were preincubated in 0.1 M PBS at room temperature. Sections were then washed with PBS and incubated with 1% Triton X-100 and 2% bovine serum albumin (BSA). For phospho-histone H3 (pH3) staining, samples were incubated with rabbit anti-pH3 (Ser10) antibody (Milipore; 1:500) under gentle shaking at 4 °C for 3 days, followed by incubation with 488 Alexa-coupled fluorescent anti-rabbit antibody (Invitrogen; 1:1000), under gentle shaking at 4 °C for 24 h. To reduce autofluorescence, sections were mounted on slides (Epredia™ SuperFrost Plus™ Adhesion slides, Thermo Scientific) and incubated with 0.3% Sudan Black in 70% ethanol. Finally, counterstaining was performed with DAPI (Mounting Medium with DAPI - Aqueous, Fluoroshield; Abcam). Slides were stored at 4 °C and protected from light.
Images corresponding to the subgranular zone (SGZ) of the hippocampal dentate gyrus were obtained with a Leica TCS-SP8X confocal microscope (oil immersion, 20× objective). Positive pH3 cells/area of the dentate gyrus (mm2) were counted in each section in at least three mice per group, and the mean of at least three sections/mouse was calculated.
2.10. Whole genome bisulfite (WGB) and RNA sequencing
DNA from 15 mice (5 CTL, 5 RES, and 5 END) was isolated using DNeasy Blood & Tissue Kit (Qiagen Inc., Ref: 69504) following the manufacturer's instructions. Proteinase K and RNase A treatments were included during the processing. Quality and quantity of DNA were assessed using Nanodrop and Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA).
Total RNA from the same 15 mice (5 CTL, 5 RES, and 5 END) was isolated using RNeasy Mini Kit (Qiagen Inc., Ref: 74104) according to the manufacturer's protocol and included DNase treatment.
Details for WGB and RNA sequencing preparation and analyses are provided in Supplementary methods and supplementary Tables.
2.11. Data availability
Paired WGB and RNA sequencing data generated for this study are available at the ENA repository (PRJEB46686 and PRJEB46807 respectively). Supplementary Tables (S1–S12) are available in the Zenodo public repository, https://doi.org/10.5281/zenodo.5513419.
3. Results
3.1. Response to the physical training interventions
To evaluate the effects of long-term resistance or endurance training on the hippocampal epigenome and transcriptome, we used wild-type mice randomly distributed into three groups: control, resistance, and endurance (Figure 1A, Figure S1A, and Table S1). Exercise intervention lasted for four weeks, after which hippocampi were resected for downstream WGBS and RNAseq analyses (Figure 1A, and Figure S1A). To determine whether the interventions were effective in terms of improvement in their respective physical capacities the mice were tested before and after the intervention. Both resistance capacity and maximal endurance capacity tests showed an enhancement in physical performance following the two training interventions (Figure 1B, Wilcoxon signed-rank test, pval < 0.05) compared with controls (Figure S1B). A more detailed analysis of each mouse separately revealed that, although significant differences were found between initial and final performance in both models of training, resistance exercise showed more interindividual variability in terms of percentage performance gain than endurance exercise (Figure 1C), something that has been described previously [39].
Figure 1.
Performance of resistance and endurance exercise in mice. A) Schema depicting the different training conditions (control, resistance, endurance) performed in this study. A total of 5 mice were used per training condition. After 4 weeks of training, the mice were sacrificed and their hippocampi resected for downstream NGS purposes. Each sample was split in order to perform paired RNAseq and WGB sequencing analyses. (See also Figure S1). B) Boxplots indicating the initial and the final performance in the context of resistance training (blue, graph on left) endurance training (yellow, graph on right). Dots denote individual mouse. The statistical significance of initial- versus final weight lifted relative to body weight (resistance) and running time (endurance, in seconds) was calculated using a Wilcoxon signed-rank test. C) Bar plot illustrating the performance gain (in percentage) for the different mice in each condition.
To determine whether the exercise intervention led to an improvement in cognitive function, the mice performed a novel object recognition test before and after the intervention. Figure S1C shows that mice from the resistance group presented an improvement in short-term memory (Wilcoxon signed-rank test, pval < 0.05), while no changes were observed in control and endurance groups, and no improvement in any group was observed with respect to long-term memory (Figure S1D). At the histological level, we found that the neurogenesis marker phosphohistone 3 (pH3) is at higher levels in trained mice, especially in the case of resistance training (Figure S1E, two-sided welch's t-test, p < 0.05).
These results show a specific improvement in the capacity trained. Both exercise interventions influenced the hippocampus, at both the functional and histological level, which was greater in those mice exposed to resistance training. In light of these results, we decided to explore whether these effects are also observed at the transcriptomic and epigenomic levels, as potential underlying mechanisms involved in the hippocampal adaptive response to training.
3.2. Physical exercise alters the DNA methylation landscape of mouse hippocampus
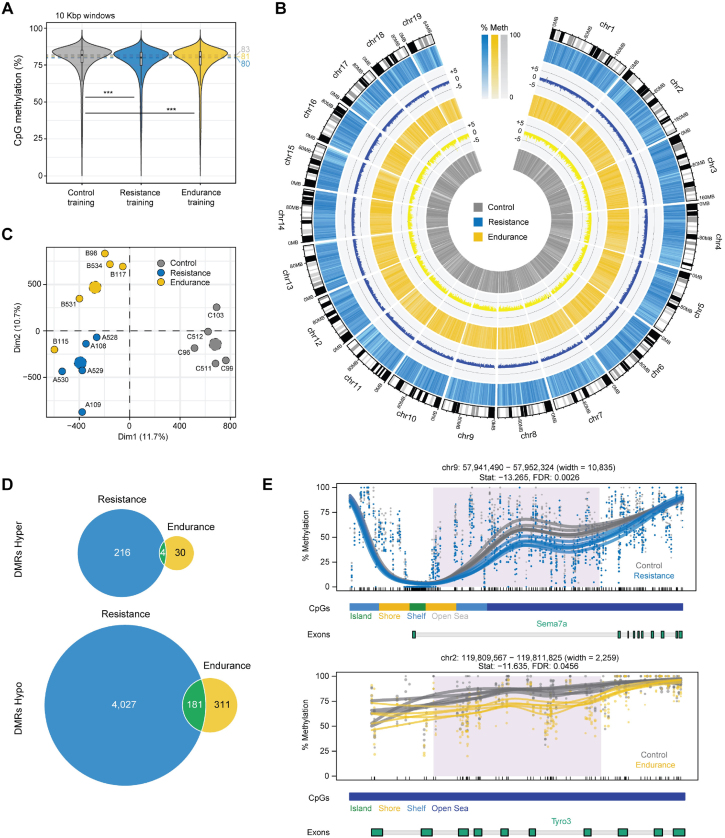
To investigate the potential implications of training on the molecular remodeling of mouse hippocampi, we initially performed WGBS experiments to carry out an in-depth characterization of the DNA methylation landscape upon physical exercise (Table S2). The mean coverage was 15.24× (13.36x-19.53×), and the mean coverage per cytosine was 10.68× (9.5x-12.98×). We observed a small but significant decrease in the overall DNA methylation status of CpG sites in the context of resistance and endurance training as compared to the control animals (Figure 2A). These changes were evenly distributed along the genome (Figure 2B) and were of sufficient magnitude to segregate the two models of exercise and the control animals into separate clusters based on a Principal Component Analysis (Figure 2C). To further explore the similarities and the differences between each training condition, we performed a differential methylation analysis by comparing the genome-wide DNA methylation status of either resistance or endurance training and the control animals. This approach revealed a total of 4,428 differentially methylated regions (DMRs) in the context of resistance and 526 DMRs for endurance training (Figure 2D) (Table S3). The number of hypomethylated loci outweighed the number of hypermethylated DMRs, as expected by the global demethylation status observed at the whole genome level. Significant examples of these epigenetic alterations are shown in Figure 2E, including the global hypomethylation of the Sema7a gene following resistance exercise and the hypomethylation of the Tyro3 gene after endurance exercise.
Figure 2.
The DNA methylation landscape of exercise stimuli in mouse hippocampus. A) Violin plots showing the global DNA methylation levels of CpG sites in mouse hippocampus subjected to different training conditions. The graph represents the percentage distribution of CpG methylation of the genome segmented in 10-Kbp genomic windows, as obtained by WGBS. The statistical significance of differences between control and either the resistance or endurance condition was calculated by means of a two-sided Wilcoxon rank sum test (∗∗∗ = p < 0.001). B) Circos-plot illustrating DNA methylation levels along the mm10 genome. CpG methylation was averaged in 10-Mbp genomic windows and the average DNA methylation value for each training condition is represented as a heatmap track. Inner bars represent overall DNA methylation changes between either the resistance (blue) or endurance (yellow) condition and the control. Scale denotes the direction and the magnitude of the change (in percentage). C) Principal component analysis for 2 × 106 most variable CpG sites across all samples included in the DNA methylation study. The proportion of variance explained for the PC1 (Dim1) and PC2 (Dim2) components is indicated. Dotted dots represent the cluster centroids D) Venn diagram depicting the total number of hyper- or hypomethylated DMRs observed in the context of resistance versus control (blue) or endurance versus control (yellow) conditions. The number of overlapping DMRs observed in both types of training is highlighted in green. E) Line plots indicating the DNA methylation profile for different DMRs and training conditions. CpG context and CpG site location are mapped to the corresponding genomic coordinates. Dots indicate the percentage of DNA methylation of individually measured CpG sites. Other statistics and DMR width are indicated at the top of the figure. Shaded areas represent locations with significantly differential methylation between exercise and control conditions as obtained by DMRseq (FDR < 0.05).
3.3. Epigenetic rewiring upon exercise involves Tet1 binding elements, pluripotency transcription factors (TFs) and is associated with DNA enhancers and repressed polycomb binding sites
To obtain more insights into the functional consequences of such epigenetic rewiring after exercise, we used a compendium of enrichment analyses at multiple levels. For a fairer statistical comparison versus a common background scenario, we first deconvoluted the DMRs into their individual CpG counterparts (dmCpGs, Figure 3A and Table S4) and performed classic CpG context and CpG location enrichment analyses. This strategy indicated that most of these CpG sites adopt the same directional change as their former DMRs (Figure S2) and that they represent a better alternative than DMRs for enrichment analysis at the genomic scale. We observed significant differences between resistance and endurance training, especially at the level of hypermethylated CpG sites (Figure 3B,C). DNA hypermethylation was particularly enriched at shelf and shore elements in the context of resistance and diminished at open sea locations (Figure 3B, Fisher's Exact Test pval < 0.001). Hypermethylated dmCpGs were more enriched at introns in the context of resistance as compared to endurance exercise and the background distribution of the WGBS experiment (Figure 3C, Fisher's Exact Test pval < 0.001). In contrast, DNA hypomethylation displayed similar distributions for both types of physical exercise and was particularly associated with intronic elements (Fisher's Exact Test p < 0.001) as compared to the background genome, suggesting that there are both common and specific alterations in the different training scenarios.
Figure 3.
Enrichment of exercise-induced hippocampal DMRs in the context of genomic location, histone marks and chromatin states. A) Graph reflecting the correspondence between DMRs and their total content of CpG sites. Values represent the total number of hyper- and hypomethylated CpGs as indicated in the figure legend. Only CpG sites with detectable DNA methylation levels in all individuals were included for downstream purposes. B and C) Stacked barplots illustrating the relative frequency of significant hyper- or hypomethylated CpGs observed in mouse hippocampus following resistance or endurance training in relation to their CpG context (B) or CpG location (C). The background distribution of all the CpGs identified in any condition of the WGBS experiment (∼18 × 106) is also indicated. D) Heatmaps showing histone mark enrichment analyses of hyper- and hypomethylated CpGs specific for resistance of endurance training as compared with the control condition. Color scales represent the magnitude of enrichment (odds ratio) of significant dmCpGs across six common histone modifications from ENCODE compared with the background distribution of all the CpGs identified in the WGBS experiment. Left legend depicts the type of mouse dataset (ENCODE) used for comparisons. E) Heatmaps illustrating chromatin state enrichment analyses of hyper- and hypomethylated CpGs in the context of resistance (top) or endurance (bottom). Color scales indicate the odds ratio of the significant dmCpGs observed across 18 chromatin states (mm10) generated from data obtained from the ENCODE consortium (see STAR methods). F) Bubble plots of enrichment of TFBS in the context of resistance or endurance conditions as determined by the information obtained from the GTRD database (mm10). Bubble color represents the statistical significance (-Log2 adjusted p-value) of TFBS enrichment in hyper- and hypomethylated CpGs respectively, as calculated using the LOLA algorithm. Dot size reflects the Log2 Odds Ratio enrichment of dmCpGs in a given TFBS dataset as compared to the background CpG distribution of the WGBS experiment. See also Figure S3.
We next performed a comprehensive region set enrichment analysis by using 6 publicly available histone datasets (H3K4me1, H3K4me3, H3K27ac, H3K27me3, H3K36me3, H3K9me3) from mouse ENCODE [40], comprising a total of 3 reference mouse brain tissues and one classical embryonic stem cell dataset. A significant enrichment of dmCpGs at more or less the same active and repressive marks was observed in both models of exercise (Figure 3D, Table S5). However, as histone marks per se do not provide the full information with respect to the final functional consequences of a given modification, we also performed an enrichment analysis based on chromatin segmentation data to explore the potential functional impact of these histone associations. We used chromHMM [41] to integrate the histone mark information from the tissues and fractionated the genome into functionally-related chromatin states as in Perez and colleagues [42]. We observed an overall enrichment of both hyper- and hypomethylated CpGs flanking transcription start sites, enhancer elements and repressed polycomb sites in the context of resistance dependent dmCpGs (Figure 3E, Table S6). On the other hand, endurance training revealed the significant enrichment of enhancer elements at hyper- and hypomethylated sites (Figure 3E).
With the aim of predicting the involvement of potential transcription factors (TFs) involved in the exercise mediated rewiring of the hippocampus epigenome, we performed a transcription factor binding site (TFBS) enrichment analysis using the Gene Transcription Regulation Database (GTRD), a comprehensive resource that includes an updated collection of uniformly processed mouse TF ChIP-seq data [43]. This analysis revealed multiple similarities between resistance and endurance training (Figure 3F and Table S7), such as a strong association between DNA hypermethylated sites and the binding sites of Jarid2, a cofactor of the Polycomb repressive complex 2 [44], which is related to cell differentiation toward neuronal lineages [45], and Onecut2, a transcription factor involved in chromatin accessibility and the induction of neural-like morphology [46]. On the other hand, the significant enrichment of Tet1 binding sites was observed at hypomethylated dmCpGs (Figure 3F) and this association was independent of the model of exercise regime employed, suggesting that the global DNA hypomethylation observed may be in part mediated by this epigenetic remodelling factor. The DNA hypomethylation scenario was strongly enriched at the putative binding sites of well-known pluripotency markers (Figure 3F), such as Nanog and Utf1 [47,48]. We confirmed these observations using an orthogonal HOMER enrichment approach (Figure S3 and Table S8). Oct and Sox pluripotency markers were enriched at DNA hypomethylated sites using the HOMER known motif database (Figure S3A). De novo prediction of putative binding motifs revealed a TFBS landscape dominated by Sox genes in the context of resistance exercise, while endurance training led to particular enrichment at Mef2 genes (Figure S3B). These differences at Mef2 associated DNA hypomethylated sites are also recapitulated in the enrichment analyses using the GTRD database (Figure 3F), indicating that despite both models of exercise displaying similar epigenetic associations, there are subtle differences that can be determinant in terms of the specific molecular rewiring that is triggered because of a given training stimuli.
3.4. Common and distinct gene expression programs observed in hippocampus following resistance or endurance training
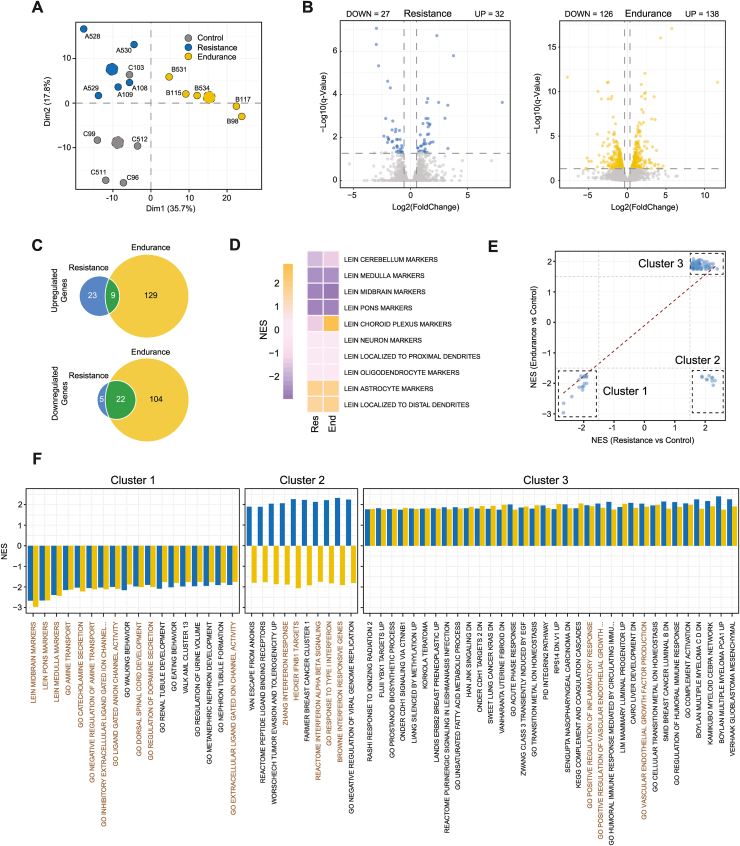
To identify the functional implications that physical exercise may exert with respect to the molecular rewiring of mouse hippocampus, we performed an in-depth transcriptional characterization on the different training intervention (Table S2). The mean number of paired reads was 48.8 M (26.8–76.4 M), and the mean percentage of mapped reads was 90.5% (84.8%–92.6%). As in the case of the DNA methylation analysis, a Principal Component Analysis using RNAseq data from paired specimens revealed the clear segregation of the exercise models from the sedentary conditions (Figure 4A). We then performed a differential gene expression analysis comparing the gene expression patterns of either resistance or endurance conditions versus the control mouse group (Table S9) (see Methods). A total of 59 differentially expressed genes (DEGs) were identified with resistance training, while endurance training revealed a total of 264 DEGs (Figure 4B). Most of the genes that were downregulated in resistance training were also downregulated in endurance exercise (Figure 4C), although the gene expression scenario was slightly different in the context of upregulated genes.
Figure 4.
Common and specific gene expression signatures of resistance and endurance stimuli. A) Principal component analysis for the 500 most variably expressed genes across all samples included in the RNA seq study. The proportion of variance explained by the PC1 (Dim1) and PC2 (Dim2) components is highlighted on the x- and y-axes respectively. Dotted dots represent the cluster centroids. B) Volcano plots illustrating the significance and the fold change of the up- or downregulated genes (colored dots) in the context of resistance (left) or endurance (right) training as compared to the control condition. C) Venn diagrams showing the total number of up- or downregulated genes observed in resistance versus control (blue) or endurance versus control (yellow) conditions. The number of overlapping DEGs observed in both types of training is highlighted in green. D) Heatmap depicting the GSEA enrichment of different exercise stimuli towards delimited brain regions denoted by the MSigDB LEIN markers datasets. Color scale indicates the calculated normalized enrichment scores (NES) for each condition as obtained from GSEA. E) Scatter plot indicating a back-to-back comparison of NES obtained from GSEA analyses of the resistance and the endurance dataset as compared to the control condition. Red dashed line indicates correlation trend between all significant GSEA datasets used for the comparison (adj P val < 0.1 in both datasets, n = 110). The three main emerging clusters are highlighted by black dashed boxes. F) Barplots reflecting the NES of the topmost significant gene sets included in the aforementioned clusters. Blue bars correspond to resistance data while yellow bars indicate NES observed for endurance exercise. See also Figure S4 and S5.
To explore the functional associations of these observed transcriptional alterations, we performed a knowledge-based approach using a Gene Set Enrichment Analysis (GSEA) [49]. We initially explored the association of the exercise conditions with particular mouse brain structures characterized by the LEIN markers dataset [50]. We observed that gene alterations in the hippocampus mediated by both resistance and endurance exercise were described as enriched at astrocyte and distal dendrites and were diminished at cerebellum and midbrain markers (Figure 4D). We found different enrichment patterns for resistance and for endurance stimuli at choroid plexus markers, which might be a consequence of the different physiological impact of each training condition. We performed a thorough GSEA analysis using the complete MSigDB collection (17,987 gene sets) [49,51] and found a total of 126 common significant gene sets from separate analyses for both models of exercise. (Table S10). According to their normalized enrichment scores in resistance and in endurance conditions, these gene sets were classified into three well differentiated clusters (Figure 4E). Cluster 1 comprised gene sets that were significantly diminished in both training models—mainly associated with midbrain, pons, and medulla markers—as well as gene ontologies related to ligand gated ion channels (Figure 4E,F). Cluster 3 contained gene sets significantly enriched in both exercise models, which was associated with vascular endothelial growth factor and the immune response. Cluster 2 was composed of gene sets with statistically significant opposing associations in resistance and in endurance exercise. This particular cluster was mainly associated with the interferon response and was significantly enriched in the context of resistance exercise (Figure 4F).
These observations were further expanded using an alternative, orthogonal modular approach. Using the genes whose variation in expression between training conditions was greatest, we were able to identify two well-differentiated gene expression modules (Figure S4A). Genes from module 1 were mainly associated with endurance stimuli, while module 2 genes were strongly positively associated with resistance exercise and diminished in the endurance condition. Ontology analyses using either classical gene ontologies or canonical pathways and Hallmark gene sets from the MSigDB collection identified distinct functional enrichments between resistance and endurance training (Figure S4B and S5). As in the case of the classical GSEA analysis, enriched canonical pathways of module 1 (endurance) displayed strong enrichment in choroid plexus markers, while module 2 was mainly characterized by significant associations with the interferon response (Figure S4B, top and S4C). Gene ontology analyses revealed the significant enrichment of synaptic signalling, dendritic spine development and glutamatergic neurons in genes from module 1, with genes from module 2 being more likely associated with the interferon response (Figure S4B, middle). At the level of hallmark gene sets, interferon was amongst the most significantly enriched pathways in the module 2 category, while module 1 displayed features found in epithelial to mesenchymal transition (Figure S4B, bottom), confirming the notion of a different functional regulatory scenario resulting from stimulation with different physical exercise regimes.
3.5. Integration of DNA methylation and gene expression data identifies robust molecular alterations in resistance or endurance mediated exercise
Integration of multiple -omic datasets often reveals molecular clues which are tightly controlled in the context of a given gene regulation program. To be able to focus on those molecular candidates displaying the most robust regulation after physical exercise, we performed a multi-level integrative analysis of DNA methylation and gene expression data. We used an enhancer linking by methylation/expression relationship (ELMER) approach using paired DNA methylation and gene expression data, as initially described by Yao and colleagues [52], see Methods). Instead of classical correlation analyses, ELMER assisted correlations were performed using the average DNA methylation value of the DMRs identified in resistance or endurance exercise versus the sedentary condition (Figure 5A) and the expression scores of their n neighboring expressed genes (5 upstream and 5 downstream of a given DMR). These analyses revealed a total of 96 and 23 robust correlations between DNA methylation and gene expression in the context of resistance and endurance conditions, respectively (Figure 5B and Table S11). Significant correlations obtained from DNA hypomethylation were associated with increased expression of neighbor target genes, while DNA hypermethylation was generally associated with gene repression (Figure 5C). We observed a little overlap between those highly correlated candidates in each exercise model. For instance, increased expression of the Krt80 gene displayed a strong correlation with the decreased methylation of a neighbouring DMR in the context of resistance, but not endurance training (Figure 5D, top). In contrast, the Ppp1r3g gene showed a strong inverse-correlation between DNA methylation and gene expression in endurance, but not in resistance (Figure 5D, bottom).
Figure 5.
Integrative analysis of DNA methylation and gene expression data reveals common and specific features of resistance and endurance training. A) Schema reflecting the number of mice (5 per group), the number of DMRs and the total number of expressed genes used for the correlations between paired DNA methylation and gene expression data. Only significant DMRs were considered for downstream purposes. B) Number of DNA methylation/gene expression correlations observed for the resistance and endurance comparisons. The graph provides separate information for hyper- or hypomethylated DMRs. C) Violin plots illustrating the Log2 fold change distribution of genes correlated with hyper- or hypomethylated DMRs in the context of resistance or endurance exercise. D) Scatterplots indicating the DNA methylation/gene expression correlation for a significantly hypomethylated DMR in resistance exercise (top) or a significantly hypomethylated DMR in the context of endurance (bottom) and their associated genes. The legend shows the Pearson's correlation score using either resistance (blue) or endurance (yellow) results versus control (gray) condition. E) Bars on the left show the correspondence between DEG-correlated DMRs and their total content of CpG sites. Values represent the total number of CpGs in these regions. These CpGs were used to reconstruct a word cloud graph with data from the TFBS enrichment analyses using the GTRD database. Word size is directly proportional to the level of enrichment in binding sites of a given transcription factor in the context of resistance (top) or endurance (bottom) exercise.
To determine the contribution of potential TFs interacting with DMRs correlated with gene-expression, we disentangled the corresponding DMRs into their individual CpG sites and we performed a TFBS enrichment analysis using the GTRD database (Figure 5E and Table S12). Tet1 was amongst the most significantly enriched TFBS in both training conditions, consistent with our previous observations using all DMR associations (Figure 3F). On the other hand, significant enrichments of Olig2 putative binding sites were observed in resistance but not endurance exercise, while, in contrast, binding sites of Kmt2b and Mef2 factors were associated with endurance but not resistance conditions, suggesting that different molecular TF networks govern the rewiring of mouse hippocampus in the two different exercise conditions.
3.6. Interleukin-1 beta (Il-1b) expression is stimulated in resistance exercise
One of the most striking differences observed between the two exercise models relies on the activation of the interferon pathway upon resistance exercise. We thus decided to focus on significant DMRs that are potentially involved in certain gene regulation programs. We found an exercise dependent DMR locus strongly associated with Il1b expression (Figure 6A). This region coincided with a developmentally activated enhancer with putative Tet1 binding sites as well as other pluripotency factors in the vicinity of this DMR (Figure 6A, right). Our integrative results indicate that Il1b cytokine is epigenetically activated, and its expression was significantly enhanced in the context of resistance but not endurance exercise (Figure 6B,C). To delve deeper into the potential consequences of Il1b activation in mouse hippocampus, we performed a computational search of potential mouse co-expressed genes in related brain and inflammation gene expression datasets using GeneMania [53]. Il1b expression interacted with multiple cytokine factors (Figure 6D), indicating a potential association between Il1b stimulation and the coordination of a complex cytokine response. We explored the overall gene expression changes in resistance exercise using the aforementioned GSEA approach and a curated Interferon signaling pathway dataset from Reactome (Figure 6E). As expected, we observed a strong significant association between this training condition and the gene targets in the Interferon Signaling dataset (Figure 6E). Resistance exercise displayed a higher interferon signature score compared to the endurance or sedentary condition, and Il1b expression displayed a positive significant correlation with the Interferon signature score (Figure 6F). This data suggests that interferon signaling enhancement is mediated by the epigenetic activation of Il1b and that this activation is solely dependent on resistance, not endurance, training conditions.
Figure 6.
Association of Il1b expression in resistance versus endurance exercise. A) Graph indicating the genomic location (mm10 coordinates) and the link between the Il1b gene and its distal associated DMR site as calculated by the ELMER algorithm. Colored tracks show the genomic location of the CpG islands and the indicated transcription factor binding sites from the GTRD database. On the right, zoom of the DMR region, including the detailed location of GTRD TFBS, the signal of the chromatin-marks H3K27ac, H3K4me3 and H3K4me1 in forebrain at different developmental timepoints, and the WGBS DNA methylation status of this region in the context of resistance or endurance exercise is also indicated. B) Boxplot depicting the gene expression differences of Il1b in mouse hippocampus between control, resistance and endurance conditions. C) Scatter plot showing the correlation between DNA methylation and gene expression of the Il1b gene. Pearson's correlation score of resistance (blue) or endurance (yellow) training versus control (grey) condition is also indicated. D) GeneMania network displaying the curated interactions between Il1b and several factors identified in the interferon signaling gene set. E) Gene set enrichment analysis showing significant links between the reactome interferon signaling gene set and the genes expressed following resistance training. F) Scatter plot indicating the correlation between the interferon related signature (x-axis) and the gene expression levels of the Il1b gene (y-axis) in the samples used in this study. The signature is strongly associated with the molecular rewiring that occurs upon resistance exercise.
4. Discussion
The benefits of physical exercise on brain and cognitive function are well-known [22,54], highlighting the importance of maintaining a healthy lifestyle not only for physical fitness, but also for correct brain functioning throughout life. To unravel the mechanisms that provide the protective effect of exercise on one of the most relevant and vulnerable brain areas, the hippocampus, we have used two murine models of long-term training: the better-known endurance training and the less explored resistance training. In this way, we have been able to analyze, for the first time, the genome-wide epigenomic and transcriptomic changes induced by either resistance or endurance training in the hippocampi of wild-type mice, allowing us to integrate these two “-omic” layers of information. This approach enabled us to determine not only those changes in hippocampus induced by exercise, but also those that are specific to the exercise model.
Our long-term resistance and endurance training protocols have been shown to be effective in terms of specific improvements in the physical capacity trained. In addition, they have also induced neurogenesis and even improved short-term memory, specifically in the case of resistance training, as we have previously described [17]. Having confirmed that both training models were able to improve these aspects of brain health, our first approach was through a genome-wide DNA methylation analysis of hippocampus samples, since epigenetics is considered the molecular link between external factors and genes [18]. Although there are several authors that have analyzed the changes in DNA methylation in the brain that are associated with exercise, these works have focused on the most studied exercise model, endurance, and on target genes [20,22,23]. Our whole genome bisulfite sequencing at single-base resolution allowed us to determine that both exercise models cause specific changes in the epigenome, mainly towards the loss of DNA methylation. A decrease in the expression pattern of several DNA methyltransferases Dnmts, the enzymes responsible for DNA methylation, was found in a study that analyzed the hippocampus of mice subjected to short-term endurance training [23], which indicates that this loss of DNA methylation might already have taken place, or started to occur, in the first week of training. On the other hand, although our results indicate that exercise is preferentially related to DNA hypomethylation regardless of the training model, we found that resistance is associated with a greater number of changes (i.e., more DMRs), which could be explained by the fact that this specific model of training actually causes more epigenetic changes in the hippocampus, or because it presents, in terms of performance, more interindividual variability than endurance exercise [39], or, indeed, it could simply be due to stochastic reasons. Future studies involving a larger number of individuals will clarify this issue.
Despite most DMRs being specific to one particular model of exercise, we did observe that differentially methylated CpGs in both conditions are associated with very similar histone marks and functionally related chromatin states. To study the potential functional role of DNA methylation changes, we analyzed those that occurred at the binding sites of transcription factors for which information was available in public repositories. As in the case of specific histone marks and chromatin functional states, we found many similarities between the two exercise models. It should be noted that in both cases there was a significant enrichment of CpGs with a loss of DNA methylation at binding sites of the demethylase Tet1, suggesting that the global DNA hypomethylation observed may be at least in part mediated by this epigenetic transcription factor. The DNA methylation changes in the Tet1 binding sites were those most significantly correlated with expression changes in each exercise model, suggesting that in addition to global DNA hypomethylation, this epigenetic transcription factor could be mediating the expression of several target genes. Although no possible molecular mechanism was found, the critical role of Tet1 in cognitive function had previously been identified through the study of the hippocampus of Tet1KO mice [55]. Our observations reaffirm the important role of this key epigenetic enzyme in the molecular rewiring of mouse hippocampus after exercise training. We also found some significant differences between exercise models with respect to hypomethylation. In fact, loss of methylation is enriched in the binding sites of pluripotency markers such as Oct and Sox in the context of resistance exercise, while in the case of endurance they were enriched in TFBS of Mef2 genes, which have been shown to be related to hippocampal learning and memory processes [56]. Globally, these results show that although both models of exercise induced a DNA hypomethylation state, exercise-associated epigenetic programs might be different in endurance and in resistance training.
At this point, certain limitations in this study should be pointed out. Despite the differences observed between resistance and endurance training, these should be considered with caution as some of them might arise from the statistical thresholds imposed by the differing number of DMRs in the initial differential methylation analysis. And secondly, consideration should be given to the potential confounding factor of the effect of specific training/learning on hippocampal gene DNA methylation and expression changes. It may be that the training/learning stimuli are stronger in the resistance training protocol than in the endurance training protocol, since the mice must be trained how to climb the ladder. These mice showed higher levels of neurogenesis.
Contrary to the results for DNA methylation, the greatest number of expression changes (i.e., DEGs) was associated with endurance exercise. When we compared the two types of exercise, we found the greatest differences in the context of upregulation, which has classically been associated with DNA hypomethylation [[57], [58], [59]]. The results of the integration analysis in the present work showed that most of the statistically significant correlations between DNA methylation and gene expression were in this sense (i.e., upregulation-hypomethylation) in both exercise models. Nevertheless, we identify potentially exercise-modified gene pathways that are specific to each exercise model. In the endurance model, gene pathways related to the regulation of synaptic transmission and plasticity, glutamatergic synapse, and the regulation of dendritic spine development are activated, all of which are compatible with adaptive neuroplasticity in the healthy brain (reviewed in [60]). We found that resistant exercise is more strongly associated with interferon response pathways. We identified that the activation of these pathways was associated with the epigenetic activation of Il1b through DNA methylation changes in a developmentally activated enhancer with putative Tet1 binding sites. Interleukin-1 beta is a pro-inflammatory cytokine that is produced by microglia in the aging brain, thus constituting a first line of defense in the maintenance of brain homeostasis. Although its chronic activation may favor neuroinflammation [61,62], interleukin-1 beta overexpression in hippocampus activates glial cells and ameliorates plaque pathology in Alzheimer's disease [63]. Although Interleukin-1 beta may be increased in certain pathophysiological conditions, it seems like this cytokine may also have beneficial effects on hippocampal synaptic plasticity, learning and memory in a concentration- and age-dependent manner [[64], [65], [66], [67], [68]]. In this sense, a recent study using a prediabetic rat model identified a significant increase in the levels of this cytokine in the hippocampus of animals subjected to both intermittent and regular exercise training, and as in our resistance exercise model, this was also associated with improved learning, memory and general cognitive function [69]. Although that work was carried out with rats and using different training procedures than those in our work [69], it reinforces the fact that exercise can have beneficial effects on memory through interferon related pathways. Future studies will determine whether the benefits of resistance training on the brain associated with interferon response pathways are due to the appropriate activation of microglia, which contributes to the maintenance of brain health.
5. Conclusion
Our results show that exercise favors DNA hypomethylation in the hippocampus, irrespective of the exercise model employed, and the status of Tet1 binding sites play an important role in transcriptional regulation. However, endurance and resistance training may activate different gene pathways, associated with neuroplasticity in the case of the former and interferon response pathways in the latter. In either case, maintaining these aspects in the brain may help to increase its resilience during aging, decreasing the negative consequences associated with this process. These genome-wide epigenomic and transcriptomic data analyzed in two different exercise models help to understand the molecular mechanisms by which exercise is a suitable preventive therapeutic strategy for brain health. Knowledge of the molecular pathways that are modulated by exercise in the brain may provide new potential therapeutic targets for future research.
Acknowledgments
The authors would like to thank Ronnie Lendrum for manuscript editing. This work was supported by the Spanish Association Against Cancer (PROYE18061FERN to M.F.F.), the Asturias Government (PCTI) co-funding 2018-2022/FEDER (IDI/2018/146 to M.F.F.), the Fundación General CSIC (0348_CIE_6_E to M.F.F.), the Health Institute Carlos III (Plan Nacional de I+D+I) co-funding FEDER (PI18/01527 to M.F.F and A.F.F.), the MINECO (DEP2015-69980-P to B.F.G.), and the Fundación Tatiana Pérez de Guzmán el Bueno (“Ayudas a Proyectos de Investigación en Neurociencia-2020” to C.T.Z and E.I.G.). R.G.U. is supported by the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER). J.R.T. is supported by a Juan de la Cierva fellowship from the Spanish Ministry of Science and Innovation MCIN/AEI /10.13039/501100011033 (IJC2018-036825-I). R.F.P. is supported by the Severo Ochoa program (BP17-114). P.P.H. is supported by Ayudas para la realización de Tesis Doctorales. Modalidad A fellowship from the University of Oviedo (PAPI-20-PF-19). We also acknowledge support from the IUOPA-ISPA-FINBA (the IUOPA is supported by the Obra Social Cajastur-Liberbank, Spain).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101398.
Contributor Information
Eduardo Iglesias-Gutiérrez, Email: iglesiaseduardo@uniovi.es.
Agustin F. Fernandez, Email: agustin.fernandez@cinn.es.
Mario F. Fraga, Email: mffraga@cinn.es.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Blair S.N., Kohl H.W., 3rd, Paffenbarger R.S., Jr., Clark D.G., Cooper K.H., Gibbons L.W. Physical fitness and all-cause mortality. A prospective study of healthy men and women. Journal of the American Medical Association. 1989;262(17):2395–2401. doi: 10.1001/jama.262.17.2395. [DOI] [PubMed] [Google Scholar]
- 2.Fiuza-Luces C., Garatachea N., Berger N.A., Lucia A. Exercise is the real polypill. Physiology. 2013;28(5):330–358. doi: 10.1152/physiol.00019.2013. [DOI] [PubMed] [Google Scholar]
- 3.Pedersen B.K., Saltin B. Exercise as medicine - evidence for prescribing exercise as therapy in 26 different chronic diseases. Scandinavian Journal of Medicine & Science in Sports. 2015;25(Suppl 3):1–72. doi: 10.1111/sms.12581. [DOI] [PubMed] [Google Scholar]
- 4.Booth F.W., Roberts C.K., Laye M.J. Lack of exercise is a major cause of chronic diseases. Comparative Physiology. 2012;2(2):1143–1211. doi: 10.1002/cphy.c110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelke K., Kemmler W., Lauber D., Beeskow C., Pintag R., Kalender W.A. Exercise maintains bone density at spine and hip EFOPS: a 3-year longitudinal study in early postmenopausal women. Osteoporosis International. 2006;17(1):133–142. doi: 10.1007/s00198-005-1938-9. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Sanles A., Sayols-Baixeras S., Castro D.E.M.M., Esteller M., Subirana I., Torres-Cuevas S., et al. Physical Activity and Genome-wide DNA Methylation: the REgistre GIroni del COR Study. Medicine & Science in Sports & Exercise. 2020;52(3):589–597. doi: 10.1249/MSS.0000000000002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes D.C., Ellefsen S., Baar K. Adaptations to endurance and strength training. Cold Spring Harbor Perspectives in Medicine. 2018;8(6) doi: 10.1101/cshperspect.a029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieman D.C., Wentz L.M. The compelling link between physical activity and the body's defense system. Journal of Sport and Health Science. 2019;8(3):201–217. doi: 10.1016/j.jshs.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nay K., Smiles W.J., Kaiser J., McAloon L.M., Loh K., Galic S., et al. Molecular mechanisms underlying the beneficial effects of exercise on brain function and neurological disorders. International Journal of Molecular Sciences. 2021;22(8) doi: 10.3390/ijms22084052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson K.I., Voss M.W., Prakash R.S., Basak C., Szabo A., Chaddock L., et al. Exercise training increases size of hippocampus and improves memory. Proceedings of the National Academy of Sciences of the U S A. 2011;108(7):3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fotuhi M., Do D., Jack C. Modifiable factors that alter the size of the hippocampus with ageing. Nature Reviews Neurology. 2012;8(4):189–202. doi: 10.1038/nrneurol.2012.27. [DOI] [PubMed] [Google Scholar]
- 12.Jackson P.A., Pialoux V., Corbett D., Drogos L., Erickson K.I., Eskes G.A., et al. Promoting brain health through exercise and diet in older adults: a physiological perspective. Journal of Physiology. 2016;594(16):4485–4498. doi: 10.1113/JP271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daviglus M.L., Plassman B.L., Pirzada A., Bell C.C., Bowen P.E., Burke J.R., et al. Risk factors and preventive interventions for Alzheimer disease: state of the science. Archives of Neurology. 2011;68(9):1185–1190. doi: 10.1001/archneurol.2011.100. [DOI] [PubMed] [Google Scholar]
- 14.ten Brinke L.F., Bolandzadeh N., Nagamatsu L.S., Hsu C.L., Davis J.C., Miran-Khan K., et al. Aerobic exercise increases hippocampal volume in older women with probable mild cognitive impairment: a 6-month randomised controlled trial. British Journal of Sports Medicine. 2015;49(4):248–254. doi: 10.1136/bjsports-2013-093184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marston K.J., Brown B.M., Rainey-Smith S.R., Peiffer J.J. Resistance exercise-induced responses in physiological factors linked with cognitive health. Journal of Alzheimer's Disease. 2019;68(1):39–64. doi: 10.3233/JAD-181079. [DOI] [PubMed] [Google Scholar]
- 16.Hawley J.A., Hargreaves M., Joyner M.J., Zierath J.R. Integrative biology of exercise. Cell. 2014;159(4):738–749. doi: 10.1016/j.cell.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Codina-Martinez H., Fernandez-Garcia B., Diez-Planelles C., Fernandez A.F., Higarza S.G., Fernandez-Sanjurjo M., et al. Autophagy is required for performance adaptive response to resistance training and exercise-induced adult neurogenesis. Scandinavian Journal of Medicine & Science in Sports. 2020;30(2):238–253. doi: 10.1111/sms.13586. [DOI] [PubMed] [Google Scholar]
- 18.Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 19.Elsner V.R., Lovatel G.A., Bertoldi K., Vanzella C., Santos F.M., Spindler C., et al. Effect of different exercise protocols on histone acetyltransferases and histone deacetylases activities in rat hippocampus. Neuroscience. 2011;192:580–587. doi: 10.1016/j.neuroscience.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 20.Trejo J.L., Herrera A., Fontán-Lozano Á., Pons S., Gradari S. Effects of physical exercise on adult hippocampal neurogenesis: changes in DNA methylation and a focus on Smad2 gene. The FASEB Journal. 2017;31(S1):lb40. [Google Scholar]
- 21.de Meireles L.C.F., Galvao F., Jr., Walker D.M., Cechinel L.R., de Souza Grefenhagen A.I., Andrade G., et al. Exercise modalities improve aversive memory and survival rate in aged rats: role of hippocampal epigenetic modifications. Molecular Neurobiology. 2019;56(12):8408–8419. doi: 10.1007/s12035-019-01675-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes J., Arida R.M., Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neuroscience & Biobehavioral Reviews. 2017;80:443–456. doi: 10.1016/j.neubiorev.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abel J.L., Rissman E.F. Running-induced epigenetic and gene expression changes in the adolescent brain. International Journal of Developmental Neuroscience. 2013;31(6):382–390. doi: 10.1016/j.ijdevneu.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovatel G.A., Elsner V.R., Bertoldi K., Vanzella C., Moyses Fdos S., Vizuete A., et al. Treadmill exercise induces age-related changes in aversive memory, neuroinflammatory and epigenetic processes in the rat hippocampus. Neurobiology of Learning and Memory. 2013;101:94–102. doi: 10.1016/j.nlm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 25.de Meireles L.C., Bertoldi K., Cechinel L.R., Schallenberger B.L., da Silva V.K., Schroder N., et al. Treadmill exercise induces selective changes in hippocampal histone acetylation during the aging process in rats. Neuroscience Letters. 2016;634:19–24. doi: 10.1016/j.neulet.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Altuna M., Urdanoz-Casado A., Sanchez-Ruiz de Gordoa J., Zelaya M.V., Labarga A., Lepesant J.M.J., et al. DNA methylation signature of human hippocampus in Alzheimer's disease is linked to neurogenesis. Clinical Epigenetics. 2019;11(1):91. doi: 10.1186/s13148-019-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanco-Luquin I., Altuna M., Sanchez-Ruiz de Gordoa J., Urdanoz-Casado A., Roldan M., Camara M., et al. PLD3 epigenetic changes in the hippocampus of Alzheimer's disease. Clinical Epigenetics. 2018;10(1):116. doi: 10.1186/s13148-018-0547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kregel K., Allen D., Booth F., Fleshner M., Henriksen E., Musch T., et al. American Physiological Society; 2006. Resource book for the design of animal exercise protocols. [Google Scholar]
- 29.Allen J.M., Berg Miller M.E., Pence B.D., Whitlock K., Nehra V., Gaskins H.R., et al. 2015. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. Journal of Applied Physiology. 1985;118(8):1059–1066. doi: 10.1152/japplphysiol.01077.2014. [DOI] [PubMed] [Google Scholar]
- 30.Conner J.D., Wolden-Hanson T., Quinn L.S. Assessment of murine exercise endurance without the use of a shock grid: an alternative to forced exercise. Journal of Visualized Experiments. 2014;90 doi: 10.3791/51846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knab A.M., Bowen R.S., Moore-Harrison T., Hamilton A.T., Turner M.J., Lightfoot J.T. Repeatability of exercise behaviors in mice. Physiology & Behavior. 2009;98(4):433–440. doi: 10.1016/j.physbeh.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayachi M., Niel R., Momken I., Billat V.L., Mille-Hamard L. Validation of a ramp running protocol for determination of the true VO2max in mice. Frontiers in Physiology. 2016;7:372. doi: 10.3389/fphys.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lira V.A., Okutsu M., Zhang M., Greene N.P., Laker R.C., Breen D.S., et al. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. The FASEB Journal. 2013;27(10):4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Deus A.P., de Oliveira C.R., Simoes R.P., Baldissera V., da Silva C.A., Rossi B.R., et al. Metabolic and cardiac autonomic effects of high-intensity resistance training protocol in Wistar rats. The Journal of Strength & Conditioning Research. 2012;26(3):618–624. doi: 10.1519/JSC.0b013e31822a5cfe. [DOI] [PubMed] [Google Scholar]
- 35.Kemi O.J., Loennechen J.P., Wisloff U., Ellingsen O. 2002. Intensity-controlled treadmill running in mice: cardiac and skeletal muscle hypertrophy. Journal of Applied Physiology. 1985;93(4):1301–1309. doi: 10.1152/japplphysiol.00231.2002. [DOI] [PubMed] [Google Scholar]
- 36.Figueiredo V.C., de Salles B.F., Trajano G.S. Volume for muscle hypertrophy and health outcomes: the most effective variable in resistance training. Sports Medicine. 2018;48(3):499–505. doi: 10.1007/s40279-017-0793-0. [DOI] [PubMed] [Google Scholar]
- 37.Gentil P., Marques V.A., Neto J.P.P., Santos A.C.G., Steele J., Fisher J., et al. Using velocity loss for monitoring resistance training effort in a real-world setting. Applied Physiology Nutrition and Metabolism. 2018;43(8):833–837. doi: 10.1139/apnm-2018-0011. [DOI] [PubMed] [Google Scholar]
- 38.Lueptow L.M. Novel object recognition test for the investigation of learning and memory in mice. Journal of Visualized Experiments. 2017;126 doi: 10.3791/55718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott B.R., Duthie G.M., Thornton H.R., Dascombe B.J. Training monitoring for resistance exercise: theory and applications. Sports Medicine. 2016;46(5):687–698. doi: 10.1007/s40279-015-0454-0. [DOI] [PubMed] [Google Scholar]
- 40.He Y., Hariharan M., Gorkin D.U., Dickel D.E., Luo C., Castanon R.G., et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature. 2020;583(7818):752–759. doi: 10.1038/s41586-020-2119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J., Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nature Protocols. 2017;12(12):2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez R.F., Tejedor J.R., Santamarina-Ojeda P., Martinez V.L., Urdinguio R.G., Villamanan L., et al. Conservation of aging and cancer epigenetic signatures across human and mouse. Molecular Biology and Evolution. 2021 doi: 10.1093/molbev/msab112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yevshin I., Sharipov R., Valeev T., Kel A., Kolpakov F. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Research. 2017;45(D1):D61–D67. doi: 10.1093/nar/gkw951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasini D., Cloos P.A., Walfridsson J., Olsson L., Bukowski J.P., Johansen J.V., et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 45.Corley M., Kroll K.L. The roles and regulation of Polycomb complexes in neural development. Cell and Tissue Research. 2015;359(1):65–85. doi: 10.1007/s00441-014-2011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Raadt J., van Gestel S.H.C., Nadif Kasri N., Albers C.A. ONECUT transcription factors induce neuronal characteristics and remodel chromatin accessibility. Nucleic Acids Research. 2019;47(11):5587–5602. doi: 10.1093/nar/gkz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Correa-Vazquez J.F., Juarez-Vicente F., Garcia-Gutierrez P., Barysch S.V., Melchior F., Garcia-Dominguez M. The Sumo proteome of proliferating and neuronal-differentiating cells reveals Utf1 among key Sumo targets involved in neurogenesis. Cell Death & Disease. 2021;12(4):305. doi: 10.1038/s41419-021-03590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Current Biology. 2011;21(1):65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 51.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao L., Shen H., Laird P.W., Farnham P.J., Berman B.P. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biology. 2015;16:105. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franz M., Rodriguez H., Lopes C., Zuberi K., Montojo J., Bader G.D., et al. GeneMANIA update 2018. Nucleic Acids Research. 2018;46(W1):W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hillman C.H., Erickson K.I., Kramer A.F. Be smart, exercise your heart: exercise effects on brain and cognition. Nature Reviews Neuroscience. 2008;9(1):58–65. doi: 10.1038/nrn2298. [DOI] [PubMed] [Google Scholar]
- 55.Rudenko A., Dawlaty M.M., Seo J., Cheng A.W., Meng J., Le T., et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79(6):1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbosa A.C., Kim M.S., Ertunc M., Adachi M., Nelson E.D., McAnally J., et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proceedings of the National Academy of Sciences of the U S A. 2008;105(27):9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calvanese V., Fernandez A.F., Urdinguio R.G., Suarez-Alvarez B., Mangas C., Perez-Garcia V., et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Research. 2012;40(1):116–131. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esteller M. Epigenetics in cancer. New England Journal of Medicine. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 59.Urdinguio R.G., Fernandez A.F., Moncada-Pazos A., Huidobro C., Rodriguez R.M., Ferrero C., et al. Immune-dependent and independent antitumor activity of GM-CSF aberrantly expressed by mouse and human colorectal tumors. Cancer Research. 2013;73(1):395–405. doi: 10.1158/0008-5472.CAN-12-0806. [DOI] [PubMed] [Google Scholar]
- 60.Mattson M.P., Arumugam T.V. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metabolism. 2018;27(6):1176–1199. doi: 10.1016/j.cmet.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J. Microglia in neurodegeneration. Nature Neuroscience. 2018;21(10):1359–1369. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X., Quan N. Microglia and CNS interleukin-1: beyond immunological concepts. Frontiers in Neurology. 2018;9:8. doi: 10.3389/fneur.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaftel S.S., Kyrkanides S., Olschowka J.A., Miller J.N., Johnson R.E., O'Banion M.K. Sustained hippocampal IL-1 beta overexpression mediates chronic neuroinflammation and ameliorates Alzheimer plaque pathology. Journal of Clinical Investigation. 2007;117(6):1595–1604. doi: 10.1172/JCI31450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ben Menachem-Zidon O., Avital A., Ben-Menahem Y., Goshen I., Kreisel T., Shmueli E.M., et al. Astrocytes support hippocampal-dependent memory and long-term potentiation via interleukin-1 signaling. Brain, Behavior, and Immunity. 2011;25(5):1008–1016. doi: 10.1016/j.bbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 65.Bourgognon J.M., Cavanagh J. The role of cytokines in modulating learning and memory and brain plasticity. Brain and Neuroscience Advances. 2020;4 doi: 10.1177/2398212820979802. 2398212820979802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hewett S.J., Jackman N.A., Claycomb R.J. Interleukin-1beta in central nervous system injury and repair. Journal of Neurodegenerative Diseases. 2012;1(2):195–211. [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider H., Pitossi F., Balschun D., Wagner A., del Rey A., Besedovsky H.O. A neuromodulatory role of interleukin-1beta in the hippocampus. Proceedings of the National Academy of Sciences of the U S A. 1998;95(13):7778–7783. doi: 10.1073/pnas.95.13.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takemiya T., Fumizawa K., Yamagata K., Iwakura Y., Kawakami M. Brain interleukin-1 facilitates learning of a water maze spatial memory task in young mice. Frontiers in Behavioral Neuroscience. 2017;11:202. doi: 10.3389/fnbeh.2017.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luvuno M., Khathi A., Mabandla M.V. The effects of exercise treatment on learning and memory ability, and cognitive performance in diet-induced prediabetes animals. Scientific Reports. 2020;10(1):15048. doi: 10.1038/s41598-020-72098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Paired WGB and RNA sequencing data generated for this study are available at the ENA repository (PRJEB46686 and PRJEB46807 respectively). Supplementary Tables (S1–S12) are available in the Zenodo public repository, https://doi.org/10.5281/zenodo.5513419.