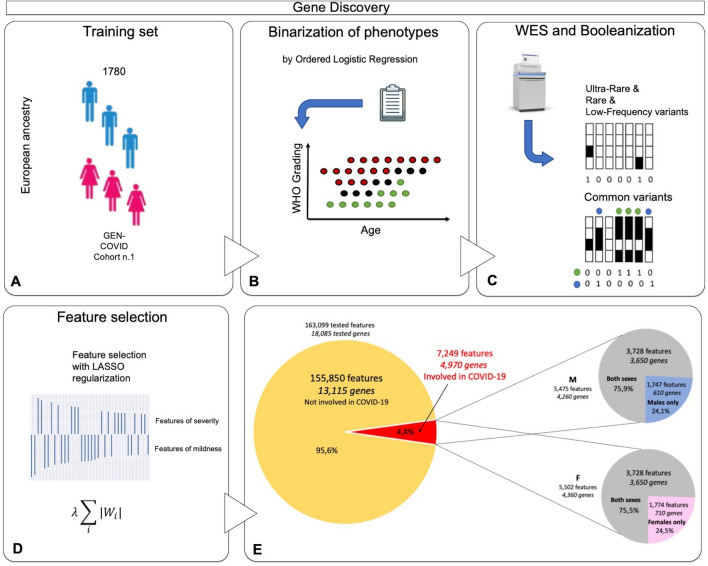
Fig. 1.
Feature selection and gene discovery. A Whole-exome sequencing (WES) data stored in the Genetic Data Repository of the GEN-COVID Multicenter Study (GCGDR) and coming from biospecimens of 1780 SARS-CoV-2 PCR-positive subjects of European ancestry of different severity were used as the training set. B Clinical severity classification into severe and mild cases was performed by Ordered Logistic Regression (OLR) starting from the WHO grading and patient age classifications. C WES data were binarized into 0 or 1 depending on the absence (0) or the presence (1) of variants (or the combination of two or more variants only for common polymorphisms) in each gene. D LASSO logistic regression feature selection methodology on multiple train-test splits of the cohort leads to the identification of the final set of features contributing to the clinical variability of COVID-19 (E). From the initial 163,099 cumulative features (divided into 36,540 ultra-rare, 23,470 rare, 13,056 low frequency and 90,033 common features) in 12 Boolean representations, the selected features contributing to COVID-19 clinical variability are 7249 and they are reported in the Supplementary Tables 3–6. The total number of genes contributing to COVID-19 clinical variability was 4260 in males and 4360 in females, 75% of which were in common