FIGURE 1.
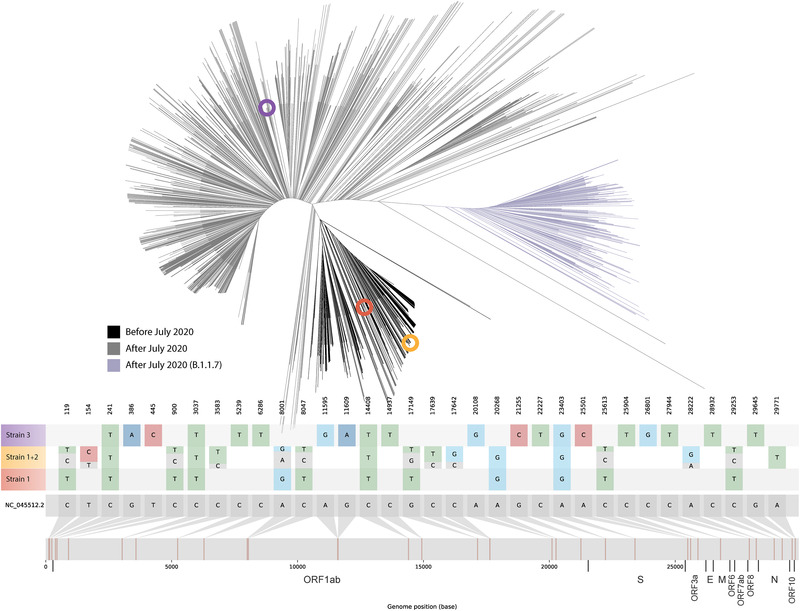
Upper panel: maximum likelihood unrooted tree with genetic distances of the three identified strains (highlighted as coloured circles within the phylogenetic tree; Strain 1 in orange, Strains 1 + 2 in ochre, and Strain 3 in purple) based on sequence alignment of 2249 positive severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) samples from cases among the Madrid population throughout the pandemic. Coloured branches are used for SARS‐CoV‐2 sequences from specimens collected during the first COVID‐19 wave (black) and after July 2020 (). The B.1.1.7 variant is shown in purple. Lower panel: location of single nucleotide polymorphisms along the SARS‐CoV‐2 reference genome for the three identified strains. Relative allele frequency of each strain (Strains 1 and 2; see also Table S1) in the superinfection event is indicated for the positions with heterozygous calls. Fasta files were deposited in GISAID (accession numbers strain 1 ‐ EPI_ISL_1547368 strain 2 ‐ EPI_ISL_1547369 and strain 3 ‐ EPI_ISL_1547363). FASTQ files were deposited at the European Nucleotide Archive (https://www.ebi.ac.uk; project reference: PRJEB47864)
