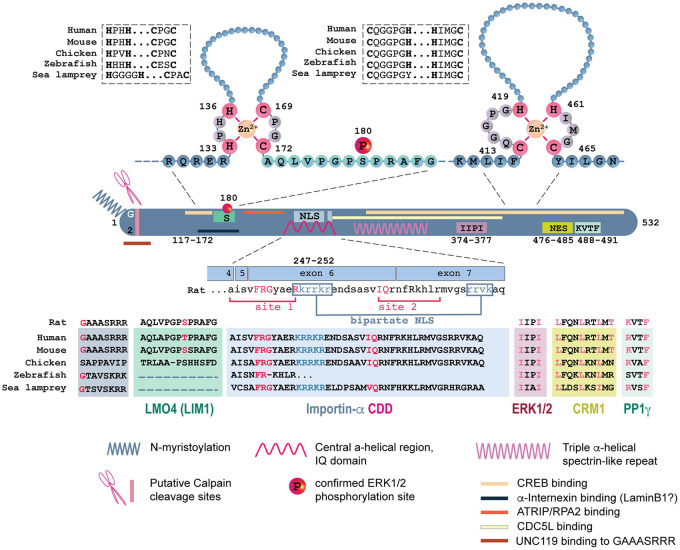
FIGURE 3.
Jacob interactome and motif conservation. Schematic representation of the binding motifs and binding regions for multiple confirmed interactors (represented by shaded boxes). The upper panel represents two well-conserved putative zinc finger domains (HH-CC and CH-HC consensus, modified from Xu et al., 2010). Cysteine or histidine residues directly binding to zinc ions are indicated in pink. Sequence conservation for the zinc finger domains is indicated in bold inside the boxes. Zebrafish Jacob (factor a) sequence is used for the panel. Amino acids encompassing the LMO4 (LIM1) binding site located directly after the first zinc finger domain are indicated in green. Color bars indicate interacting partners. Color boxes indicate motifs. Crucial amino acids residues within the motives are indicated in red. RPA2, replication protein A 2; ATR, ataxia telangiectasia and Rad3-related protein; CDC5L, Cell Division Cycle 5 Like; ATRIP, ATR interacting protein; CDD, Caldendrin; UNC119, solubilizing factor.