Highlights
-
•
The metabolomic workflow for disease biomarker discovery was optimized.
-
•
Two separation modes using T3 and amide columns and two polarity modes were used.
-
•
Metabolome and lipidome coverage was improved via an integrative workflow.
-
•
Increased lysophosphatidylcholines, diacylglycerols and acylcarnitines were noted in sera of NAFLD patients.
Abbreviations: ACN, acetonitrile; amide-pos, amide column-based HILIC separation plus the positive ion mode; amide-neg, amide column-based HILIC separation plus the negative ion mode; DGs, diacylglycerols; EICs, extracted ion chromatograms; ESI− and ESI+, ionization polarity modes; FA, Formic acid; FC, fold change; HCC, hepatocellular carcinoma; HFD, high-fat diet; HILIC, hydrophilic interaction chromatography; LPC, lysophosphatidylcholine; MCD, methionine-choline-deficient; LE, Leucine enkephalin; MGs, monoacylglycerols; MS, mass spectrometry; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; OPLS-DA, orthogonal partial least square discriminant analysis; PCs, phosphorylcholines; PEs, phosphatidylethanolamines; PKC∊, protein kinase C∊; ROC, receiver operating characteristic; RPLC, reversed-phase liquid chromatography; T3-neg, T3 column-based reverse phase separation plus the negative ion mode; T3-pos, T3 column-based reverse phase separation plus the positive ion mode; TIC, total ion chromatogram; VIP, variable importance
Keywords: NAFLD, Metabolic biomarkers, Lipids, Metabolomics, Acylcarnitines
Abstract
Background
The worldwide prevalence of non-alcoholic fatty liver disease (NAFLD) has stimulated work to identify biomarkers and develop effective treatments. Metabolomics is an emerging tool that has been widely applied to discover biomarkers and simultaneously uncover pathological mechanisms. Here, we aim to optimize metabolomic acquisition with the goal of obtaining a systemic metabolic profile to unravel the potential link between dysregulated metabolism and NAFLD.
Methods
We analyzed serum samples collected from healthy subjects (n = 8) and NAFLD patients (n = 8) via an integrative analytical workflow using two orthogonal separation modes with T3 and amide columns and two ionization polarity modes on a UPLC-ESI-Q/TOF. Data dependent acquisition was employed for data acquisition. Differentially expressed metabolites and lipids were identified by comparing the collected metabolic and lipidomic profiles between the healthy subjects and NAFLD patients.
Results
The integrative LC-MS/MS analytical workflow employed here features an improved coverage of metabolites and lipids, which leads to the identification of 20 potential biomarkers of NAFLD, including lipids, acylcarnitines, and organic acids.
Conclusions
This pilot study has identified potential biomarkers for NAFLD and revealed corresponding dysregulated metabolic pathways related to NAFLD's occurrence and progression, establishing a molecular basis for NAFLD diagnosis and therapeutic intervention.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a clinicopathologic syndrome. It is usually caused by excessive fat deposition in hepatocytes. NAFLD has been associated with diabetes, hypertension, dyslipidemia, and obesity [1]. It covers a broad spectrum of hepatopathy ranging from simple steatosis to non-alcoholic steatohepatitis (NASH) with varying degrees of fibrosis. It can develop into cirrhosis and, ultimately, hepatocellular carcinoma (HCC) [2]. Currently, NAFLD has become the most common chronic liver disease in developed countries and affects about a quarter of the world's population [3].
Hepatic ultrasonography and biopsy [4], [5] are the conventional detection approaches for definitive diagnosis. However, liver biopsy is an invasive, painful, and costly procedure [6]. In contrast, ultrasonography can obtain liver histomorphology images that indicate the severity and disease stages for NAFLD. Nevertheless, both measures cannot provide clues regarding the underlying mechanisms of NAFLD occurrence and progression, and, subsequently, suggest pharmacological interventions [7]. Therefore, a broad, non-invasive medical method that can provide a molecular basis for NAFLD diagnosis and therapeutic intervention is urgently needed.
Metabolomics is a relatively new field that complements genomics, transcriptomics, and proteomics. It collects comprehensive information on identities and dynamic abundance levels of metabolites and lipids in cells, tissues, organisms, and body fluids, allowing the obtainment of an overall metabolic profile from different subjects [8]. Thus, metabolomics studies that aim to identify potential therapeutic biomarkers for diseases are emerging [9], [10]. Specifically, metabolomics has been applied to characterize various biomarkers for NAFLD. For instance, in a high-fat diet (HFD)-induced NAFLD mouse model, 30 potential biomarkers including serum glucose, total cholesterol, and hepatic triglyceride were identified, and their abundance was found to be intimately associated with the development of NAFLD [11]. Besides animal models, mass spectrometry (MS)-empowered metabolomics has been clinically applied to identify various metabolite and lipid biomarkers to distinguish NASH and steatosis patients, and reveal candidates, such as glycerophospholipids, sphingolipids, and sterols, as potential NAFLD biomarkers [5]. Moreover, additional LC-MS/MS-based metabolomics studies reported elevated sphingomyelins and phosphatidylcholine in the serum of NAFLD patients when compared to healthy controls [12]. Collectively, these studies demonstrate that metabolism is significantly dysregulated between the NAFLD and control groups, hence, identifying the differential metabolic and lipidomic markers are anticipated to advance clinical diagnosis and inspire the development of treatment for NAFLD patients.
Nevertheless, these non-targeted metabolomics studies that aim to characterize the biomarkers for NALFD usually only employ one specific separation mode and one specific polarity mode for data acquisition. Thus, it is almost impossible to achieve comprehensive retention of all hydrophilic and hydrophobic substances by solely employing either hydrophilic interaction chromatography (HILIC) or reversed-phase liquid chromatography (RPLC). Similarly, only using positive ion mode or negative ion mode cannot obtain all substances with different ionization characteristics. Given the complementarity between distinct chromatographic columns and ionization modes, we have attempted to combine different chromatographic separation columns and ionization modes to optimize the metabolome coverage for biomarker discovery.
We designed the metabolome study by integrating four analytical modes, including the T3 column-based reverse phase separation plus the positive ion mode (T3-pos), the T3 column-based reverse phase separation plus the negative ion mode (T3-neg), the amide column-based HILIC separation plus the positive ion mode (amide-pos), and the amide column-based HILIC separation plus the negative ion mode (amide-neg). Database search-assisted metabolic identification shows that each of the four modes is capable of detecting unique metabolites that cannot be addressed by the other analytical modes. Accordingly, the optimal metabolome coverage should be able to be achieved by the integration of the four modes. A total of 20 potential metabolic biomarkers associated with NAFLD were identified by the integrative analytical mode using a limited number of serum samples collected from clinical NAFLD patients and healthy subjects in this pilot study. Notably, our metabolomic analysis revealed the significantly elevated concentration of selected lipids, acylcarnitines, and organic acids in NAFLD patients. By analyzing the small pool of patients recruited for this pilot study, we have demonstrated that metabolome coverage improvement can be achieved using the integrative analytical modes (i.e., T3-pos, T3-neg, amide-pos, amide-neg), whereas a single analytical method was not adequate for comprehensive coverage of metabolomic/lipidomic biomarkers. We anticipate that the integrative analytical mode we tested for biomarker discovery will contribute to translational research in future metabolomics research.
Material and methods
Patients
We recruited NAFLD patients from the Nanjing First Hospital (Nanjing, Jiangsu Province, China). Eight subjects with NAFLD and eight healthy controls participated in the metabolomic study. Hepatic ultrasonography was performed on all human subjects by the same investigator to confirm the presence or absence of steatosis. None of the patients were on any medications known to cause hepatic steatosis, nor were they taking vitamin supplements. Dietary advice and standardized overnight fasting were implemented to ensure the collected human serum samples' uniformity and consistency. All participants provided informed consent, and the local ethics committee approved the protocol of this study. Fasting blood samples were drawn via venipuncture by clinical nurses. The blood sample was collected and then centrifuged at 3,500 g for 10 min at 4 °C, and the clear serum was stored at − 80 °C.
Reagents
Formic acid (FA), ammonia solution, and ammonium formate were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Ultra-pure water was obtained from the Milli-Q Pure Water System (Bedford, MA, USA). LC-MS grade methanol and acetonitrile (ACN) were purchased from Merck (Darmstadt, Germany).
Sample preparation for metabolomic analysis
Frozen human serum samples were thawed at 4 ℃ and processed for metabolomic analysis as previously reported. Briefly, metabolites were extracted by adding 400 μL of ice-cold methanol to a 100 μL aliquot of human serum followed by 2 min vortexing. The mixture was then centrifuged at 4 °C and 15,000 g for 5 min, and the supernatant was collected. The remaining pellet was repetitively extracted with ice-cold methanol. All the supernatants were pooled and evaporated to dryness. The extracts were subsequently reconstituted in 100 μL of 50% methanol containing 0.1% FA, centrifuged at 18,000 g for 10 min, and then collected. For quality control, aliquots of each sample were pooled and injected throughout the injection queue to monitor the reproducibility of instrumental performance.
Chromatography
Chromatographic separation of serum metabolites was performed on a UPLC system (I Class Plus, Waters, Milford, MA, USA). Two chromatographic separation modes were employed for improved metabolome coverage using an ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm, Waters) for RP separation and an XBridge BEH Amide column (4.6 × 100 mm, 3.5 μm, Waters) for HILIC mode separation. During separation, the T3 column temperature was set at 40 ℃, and the flow rate was 0.4 mL/min. Mobile phase A consisted of 95 % 10 mM ammonium formate buffer, and mobile phase B consisted of ACN. The gradient was set as follows: 0–1.00 min, 0% B; 1.00–4.00 min, 0–35% B; 4.00–15.50 min, 35–95% B; 15.50–18.00 min, 95% B; 18.10–23.00 min, 0% B. The temperature of the BEH amide column was set at 40 ℃, and the flow rate was 0.4 mL/min. Mobile phase A consisted of 95 % 10 mM ammonium acetate buffer and 5% ACN (pH adjusted to 9), and mobile phase B consisted of ACN. The gradient was set as follows: 0–3 min, 85 % B; 3–6 min, 85–30% B; 6–15 min, 30–2% B; 15–18 min, 2% B; 18–19 min, 2–85% B; and 19–26 min, 85% B.
Mass spectrometry
Metabolites and lipids were detected on a UPLC-QTOF-MS/MS mass spectrometer (Waters, Milford, MA, USA) equipped with both ionization polarity modes (ESI − and ESI + ). The detection parameters were set based on previous literature as follows: ion spray voltage at + 3.0 kV (ESI + ) and − 2.5 kV (ESI − ), sampling cone at 40, source offset at 80, source temperature at 100 ℃, desolvation temperature at 450 ℃, cone gas at 50 L/h, desolvation gas at 600 L/h. MS parameters were set as follows: MS scan m/z range at 50–700 Da, MS survey scan time at 0.20 s, MS/MS scan time at 0.15 s, MS survey switching threshold at 8000, MS/MS collision energy at 10–20 V for the low-end and 24–50 V for the high-end. All acquisition was accompanied by lockspray infusion of 200 ng/mL leucine encephalin (m/z 556.2771 at positive ion mode and m/z 554.2615 at negative ion mode). Metabolomics data acquisition was conducted using the software MassLynx 4.1 (Waters).
Data processing
The control and NAFLD subjects’ metabolomics and lipidomics profiles of serum samples were compared using Progenesis QI (version 2.0, Waters) for potential biomarker discovery. The metabolic and lipidomic identification process in Progenesis QI involved the following steps: create a new experiment, import data, review alignment, experiment design setup, peak picking, review deconvolution, identify compounds, review compounds, and compound statistics. Specifically, [M + H]+, [M + Na]+, [M + K]+, [M + NH4]+, [M + H-H2O]+ were set for possible positive adducts and [M−H]-, [M−H2O−H]-, [M + FA-H]- were selected for possible negative adducts in creating a new experiment. Leucine enkephalin (LE) lock mass m/z 556.2766 was set in positive mode, and m/z 554.2620 was set in the negative mode. For compound identification, MS and MS/MS fragment ions generated from compounds of interest were searched against those documented in databases including the Human Metabolome Database (http://www.hmdb.ca/), MONA (http://mona.fiehnlab.ucdavis.edu/), and METLIN (https://metlin.scripps.edu/). The identification criteria were set as follows: a mass error of 10 ppm was allowed for precursor ion matching and 20 ppm for fragment ion matching; the error of isotope distribution was set as<20%; the library score of precursor ions must be higher than 40 and the fragmentation spectrum of each putative biomarker was manually inspected to verify the assigned identity. When comparing the abundance levels of lipids/metabolites between the two groups for biomarker discovery, the abundance of each assigned compound was calculated by integrating the corresponding chromatographic peak followed by normalization against the total ion chromatogram (TIC) of each sample using Progenesis QI. Manual inspection of extracted ion chromatograms (EICs) of each identified compound was also performed to ensure data integrity.
Statistical analysis
The metabolomics data were processed by QI and further exported to EZinfo software (version 3.0, Waters) for orthogonal partial least square discriminant analysis (OPLS-DA). Upon OPLS-DA, compounds with variable importance in the projection calculated based on S-plots (VIP) > 1, P-value < 0.05 (Student’s t-test) was considered statistically significant difference and fold change (FC) > 1.5 were summarized and assigned as a potential biomarker of NAFLD. Specifically, for identical compounds yet detected in different analytical modes (chromatography and/or ion polarity), we adopted the peak displaying the smallest FC for further data analysis. After metabolic identification, bioinformatic analysis of assigned metabolomic and lipidomic identities, including pathway analysis and receiver operating characteristic (ROC) curve were plotted on MetaboAnalyst 3.0 (http://www.metaboanalyst.ca/).
Results and discussion
Metabolomics analysis of NAFLD using UPLCQ-TOF/MS
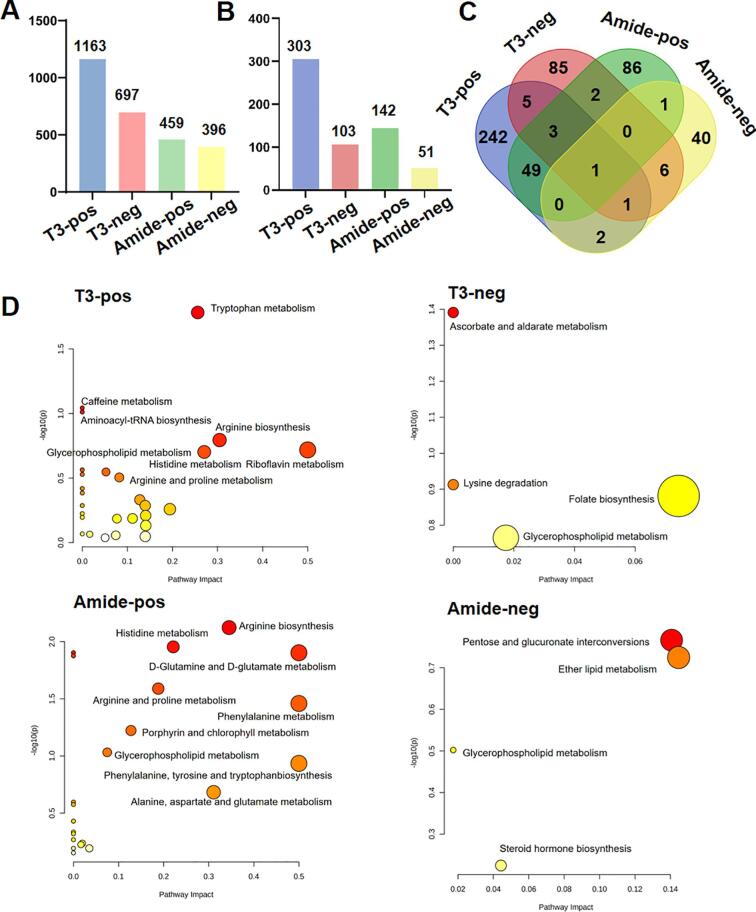
Chromatographic separation and MS acquisition conditions are of paramount importance for comprehensively detecting metabolic and lipidomic components in biological samples. Therefore, different chromatographic columns and MS acquisition modes were combinatorialy employed to achieve increased detection coverage compared to single separation and MS polarity conditions. The integration of the HSS T3 RP column with a BEH amide column and the use of both positive and negative MS acquisition modes allows for detection of hydrophilic and hydrophobic metabolites. Specifically, we extracted characteristic peaks from the serum samples using the T3 column and amide column in both positive and negative modes. Of the extracted peaks, 1163, 697, 459, and 396 compounds were detected when T3-pos, T3-neg, amide-pos, and amide-neg mode were employed, respectively (Fig. 1A). Subsequently, metabolic identification was tentatively made by searching these compounds against MS and MS/MS spectra in public databases exemplified by METLIN, MONA, and HMDB. Under the matching scores threshold of 40, we retrieved 303, 103, 142, and 51 MS and MS/MS metabolites as high-confidence identifications by using the four analytical modes, respectively (Fig. 1B).
Fig. 1.
Identification of metabolites and pathway analysis through different acquisition modes. (A) Histogram showing the distribution of the detected compounds among the T3-pos, T3-neg, amide-pos, and amide-neg analytical modes. (B) Histogram showing the distribution of the identified compounds among the T3-pos, T3-neg, amide-pos, and amide-neg analytical modes. (C) Venn diagram showing the distribution of the identified compounds among the T3-pos, T3-neg, amide-pos, and amide-neg analytical modes. (D) Metabolic pathways involving the identified metabolites in the control and NAFLD subjects were collected by the T3-pos, T3-neg, amide-pos, and amide-neg analytical modes. The larger the representative circle's size and the deeper the red color intensity, the greater the impact on the respective pathway. A total of 16 serum samples from NAFLD patients and healthy controls were acquired on LC-Q/TOF under different analytical modes, and datasets acquired under different analytical modes were subjected to data analysis and comparison. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we asked whether the integration of the four analytical modes truly benefits the detection coverage. Notably, for high-confidence metabolites detected by the T3 column-based chromatographic separation in the positive and negative ion modes, a total of 10 compounds were shared between the T3-pos and T3-neg modes, with 293 metabolites being exclusively detected in the T3-pos mode vs. 93 in the T3-neg mode (Fig. 1C). Only 2 compounds were shared between the amide-pos and amide-neg data, with 140 metabolites being exclusively detected in the amide-pos mode vs. 49 in the amide-neg mode. Our results highlight the necessity of conducting metabolomic analysis in both positive and negative ionization modes.
We next explored whether chromatographic behaviour would markedly impact the detection coverage for metabolites of similar ionization properties. We found 53 overlapping metabolites were detected in both T3-pos and amide-pos modes, whereas 242 metabolites were only detected in the T3-pos mode and 86 metabolites were exclusively identified in the amide-pos mode. The analysis of T3-neg and amide-neg data has led us to assign 8 overlapping metabolites in both analytical modes. In contrast, only 85 metabolites were exclusively detected in the T3-neg mode and 40 metabolites were identified in the amide-neg mode (Fig. 1C).
We then asked whether the metabolites detected by different analytical modes belong to distinct metabolic pathways (Fig. 1D). We found metabolites detected in the T3-pos mode are involved in tryptophan metabolism, arginine biosynthesis, caffeine metabolism, aminoacyl-tRNA biosynthesis, and riboflavin metabolism. In contrast, metabolites detected using the same column, but in the negative mode, are involved in the ascorbate and alternate metabolism and lysine degradation. The enriched pathways under the amide-pos mode include arginine biosynthesis, histidine metabolism, D-glutamine, D-glutamate metabolism, arginine, and proline phenylalanine metabolism, and porphyrin and chlorophyll metabolism. Metabolic pathways including pentose and glucuronate interconversions and lipid metabolism pathways were enriched in the amide-neg analytical mode. Collectively, we summarized that it is necessary to employ different chromatography separation and polarity modes for achieving comprehensive metabolome and lipidome coverage due to their variant physicochemical properties. The improved coverage is anticipated to allow us to glean information regarding the altered biological pathways in response to physiological and pathological changes.
Metabolic and lipidomic changes in NAFLD patients sera
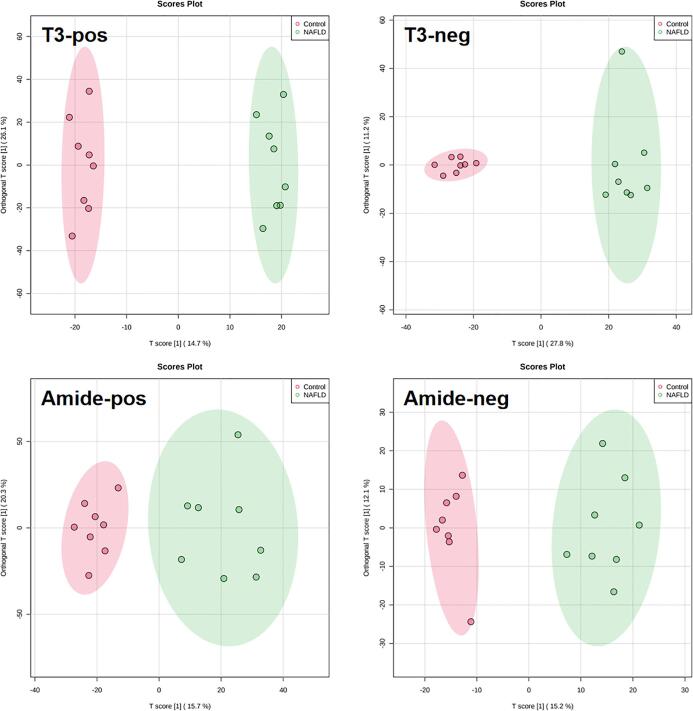
We then employed the four analytical modes to obtain a holistic profile from the sera of NAFLD patients and healthy subjects. As shown in Fig. 2, we found the Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) plots showed that the healthy subjects and NAFLD patients all exhibited different patterns under the four analytical modes (R2X = 98% Q2 = 97% for the T3-pos mode, R2X = 99% Q2 = 99% for the T3-neg mode, R2X = 99% Q2 = 98% for the amide-pos mode, R2X = 99% Q2 = 98% for the amide-neg mode). These scoring plots signify the distinct metabolic and lipidomic patterns between NAFLD and healthy subjects.
Fig. 2.
OPLS-DA score plots derived from the serum metabolomics datasets collected from the control and NAFLD subjects using LC-Q/TOF via the T3-pos, T3-neg, amide-pos, and amide-neg analytical mode (n = 8/group).
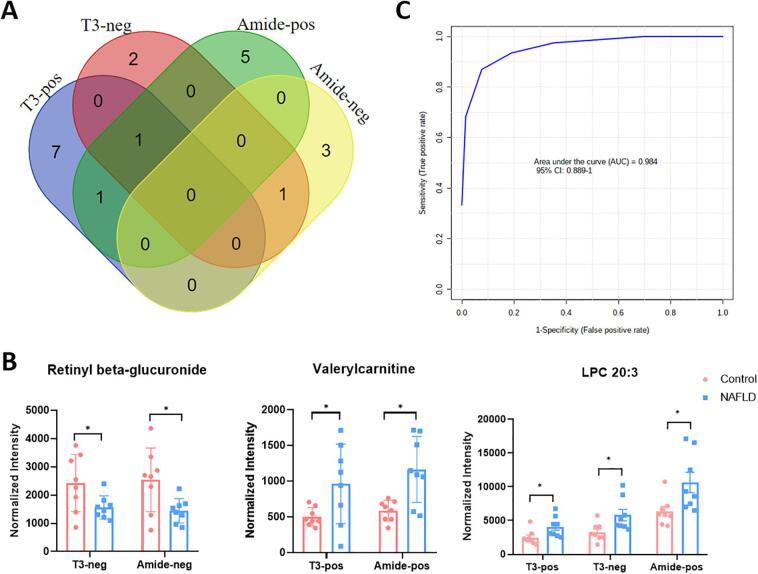
To elaborate on which metabolites and lipids show significantly aberrant levels in NAFLD patients, a student's t-test followed by Tukey's test for post-hoc (p value < 0.05) was performed for metabolites with VIP > 1 based on the established OPLS-DA models. An FC cutoff of 1.5 was further calculated to identify the potential metabolic and lipidomic biomarkers that display differentiation in NAFLD patients and healthy subjects. A total of 20 potential metabolic biomarkers were combinatorially identified using the T3-pos, T3-neg, amide-pos, and amide-neg analytical modes (summarized in Table 1), and their distribution in each analytical mode is illustrated in Fig. 3A. Specifically, 9, 4, 7, and 4 putative metabolic biomarkers were detected in the T3-pos, T3-neg, amide-pos, and amide-neg modes, respectively. Specifically, lysophosphatidylcholine (LPC) 20:3 was detected in T3-pos, T3-neg, and amide-pos modes, whereas valerylcarnitine was detected in both T3-pos and amide-pos modes, and retinyl-beta glucuronide was detected in both T3-neg and amide-neg modes (Fig. 3B). These findings confirmed the necessity of adopting multiple chromatographic separation and polarity modes for metabolic biomarker discovery. Expectedly, we found that metabolites detected in both analytical modes exhibited consistent trends in abundance changes.
Table 1.
Potential metabolic biomarkers differentiating the NAFLD and control subjects using different analytical modes (T3-pos, T3-neg, amide-pos, amide-neg). All metabolic identifications have been confirmed by MS and MS/MS. Fold change was calculated by comparing the abundance level of the metabolite in the NAFLD group vs. the control group (NAFLD/Control). P value was calculated using the Student’s t-test.
| Mode | Metabolite | Formula | Mass (Da) | Retention time (min) | Adductsa | Observed (m/z) | Mass error (ppm) | Fold change | P value |
|---|---|---|---|---|---|---|---|---|---|
| T3-pos | Valerylcarnitine | C12H23NO4 | 245.1627 | 3.31 | [M + H]+ | 246.1694 | −2.25 | 1.92 | 0.0386 |
| Octanoylcarnitine | C15H29NO4 | 287.2097 | 4.47 | [M + H]+ | 288.2164 | −1.74 | 4.65 | 0.0124 | |
| Hydroxyundecanoyl carnitine | C18H35NO5 | 345.2515 | 5.08 | [M + H-H2O]+ | 328.2480 | −0.59 | 2.01 | 0.0197 | |
| Hydroxydodecanedioic acid | C12H22O5 | 246.1461 | 4.50 | [M + H-H2O]+ | 229.1428 | −2.39 | 2.11 | 0.0045 | |
| PE(17:0/0:0) | C22H46NO7P | 467.3018 | 7.40 | [M + H]+ | 468.3091 | 1.26 | 1.76 | 0.0121 | |
| LPC(18:3) | C26H48NO7P | 517.3168 | 7.81 | [M + H]+ | 518.3242 | 0.22 | 1.55 | 0.0123 | |
| LPC(20:3) | C28H52NO7P | 545.3481 | 8.91 | [M + H]+ | 546.3561 | 1.27 | 1.64 | 0.0300 | |
| 25-Hydroxyvitamin D3-26,23-lactol | C27H42O4 | 430.3083 | 9.38 | [M + H]+ | 431.3155 | −0.20 | 1.60 | 0.0203 | |
| MG(18:1) | C21H40O4 | 356.2925 | 13.77 | [M + H]+ | 357.3016 | −0.53 | 3.65 | 0.0250 | |
| T3-neg | Deoxycholic acid 3-glucuronide | C30H48O10 | 568.3247 | 5.83 | [M−H]- | 567.3165 | −1.70 | 4.76 | 0.0084 |
| Tuftsin | C21H40N8O6 | 500.3071 | 5.34 | [M−H]- | 499.3007 | 1.75 | 3.89 | 0.0211 | |
| LPC(20:3) | C28H52NO7P | 545.3481 | 8.91 | [M + FA-H]- | 590.3411 | −9.61 | 1.77 | 0.0210 | |
| Retinyl beta-glucuronide | C26H38O7 | 462.2618 | 5.45 | [M−H]- | 461.2554 | 1.93 | 0.64 | 0.0427 | |
| Amide-pos | LPC(14:0) | C22H46NO7P | 467.3012 | 3.39 | [M + H]+ | 468.3097 | 2.65 | 2.73 | 0.0026 |
| DG(20:3/20:4) | C43H70O5 | 666.5234 | 2.43 | [M + H]+ | 684.5542 | 1.62 | 7.22 | 0.0261 | |
| Valerylcarnitine | C12H23NO4 | 245.1627 | 7.36 | [M + H]+ | 246.1697 | −0.99 | 1.99 | 0.0045 | |
| 3-Hydroxy-cis-5-tetradecenoylcarnitine | C21H39NO5 | 385.2828 | 3.34 | [M + H-H2O]+ | 368.2798 | 0.58 | 1.69 | 0.0057 | |
| LPC(20:3) | C28H52NO7P | 545.3491 | 3.31 | [M + H]+ | 546.3564 | 1.83 | 1.67 | 0.0224 | |
| Hydroxylauroylcarnitine | C19H37NO5 | 359.2672 | 5.40 | [M + H-H2O]+ | 342.2641 | 0.69 | 1.55 | 0.0251 | |
| DG(18:1)/18:2) | C39H70O5 | 618.5191 | 2.45 | [M + H-H2O]+ | 636.5558 | −5.16 | 2.74 | 0.0423 | |
| Amide-neg | Cortolone-3-glucuronide | C27H42O11 | 542.2727 | 7.33 | [M−H]- | 541.2697 | 7.93 | 2.58 | 0.0004 |
| Tetrahydroaldosterone-3-glucuronide | C27H40O11 | 540.2571 | 7.22 | [M−H]- | 539.2548 | 9.24 | 2.02 | 0.0007 | |
| LPC(18:0) | C26H54NO7P | 523.3638 | 3.34 | [M + FA-H]- | 568.3603 | −3.31 | 2.13 | 0.0323 | |
| Retinyl beta-glucuronide | C26H38O7 | 462.2618 | 3.11 | [M−H]- | 461.2564 | 4.17 | 0.57 | 0.0228 |
The metabolite adducts of the highest intensity were chosen to represent the abundance of given metabolites.
Fig. 3.
Identification of potential metabolic biomarkers under different acquisition modes. (A) Venn diagram shows the potential biomarkers' distribution among the T3-pos, T3-neg, amide-pos, and amide-neg modes between NAFLD and control subjects. (B) Abundance changes of retinyl beta-glucuronide, valerylcarnitine, and LPC (20:3) in the NAFLD and control subjects collected in different modes. Bar plot represents the normalized intensity (normalized by total ion chromatogram, TICs) of selected metabolites in serum. Data are expressed as normalized intensity ± SEM (n = 8/group), and fold change is indicated as FC. A comparison was made between NAFLD and control subjects using the Student's t-test. *, significantly different between the control and NAFLD subjects at p < 0.05; **, significantly different between the control and NAFLD subjects at p < 0.01. (C) ROC curve analysis of the potential biomarkers for NAFLD.
ROC curve analysis of the potential biomarkers
We then created ROC curves to explore whether these putative biomarkers have potential for clinically diagnosing NAFLD. The metabolic biomarkers identified to differentiate serum profiles collected from the healthy and NAFLD subjects were analyzed using binary logistic regression on Metaboanalyst to obtain the predictive probability. Although the patient cohort was small and there may have been overfitting, the AUC of putative biomarkers reached 0.984 (Fig. 3C), which appears to indicate that the identified metabolites can differentiate healthy subjects from NAFLD patients based on serum metabolic and lipidomic profiles. However, we also realize that the small size of the cohort enrolled in this study could have easily resulted in overfitting of the model. As with most pilot studies, it is clear that a large population of healthy subjects and NAFLD patients will be necessary to verify the diagnostic value of these putatively assigned biomarkers of NAFLD.
Metabolomic and lipidomic biomarkers of NAFLD and their pathological roles
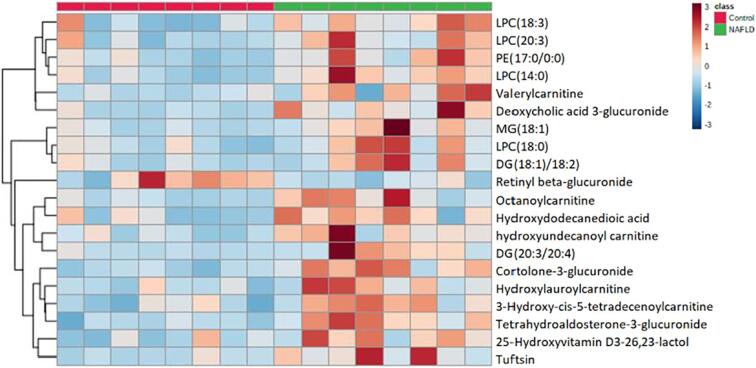
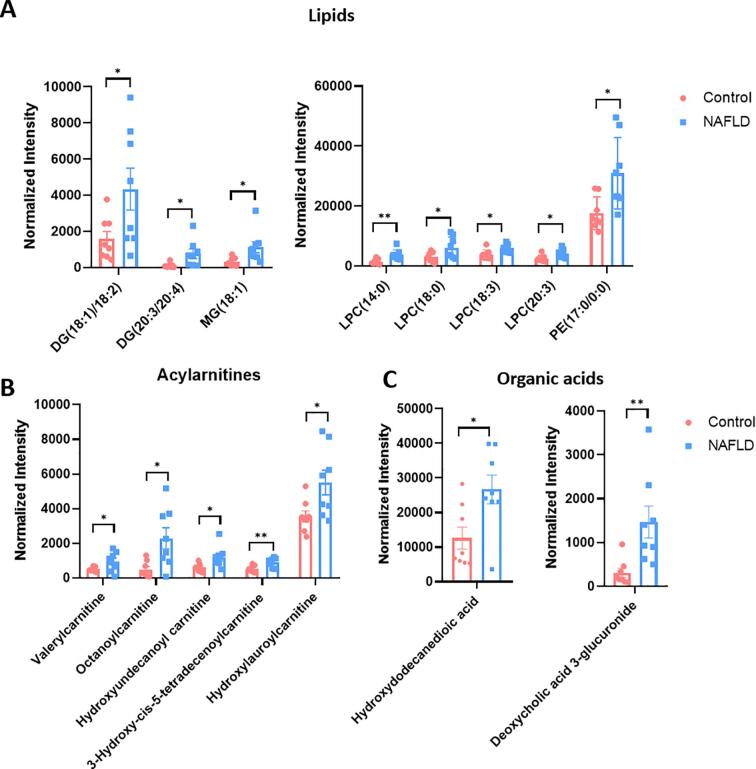
We created a heatmap to visualize the 20 potential biomarkers consisting of lipids, acylcarnitines, and organic acids that we used to differentiate between NAFLD and healthy subjects (Fig. 4). PE(17:0/0:0), LPC 14:0, LPC 18:0, LPC 20:3, LPC 18:3, DG(18:1/18:2), DG(20:3/20:4), and MG (18:1) all displayed increased abundance in the NAFLD group as shown in Fig. 5A. These lipids can be classified as phosphatidylethanolamines (PEs), phosphorylcholines (PCs), lysophosphatidylcholines (LPCs), diacylglycerols (DGs), and monoacylglycerols (MGs). Indeed, the increased PCs and PEs in NAFLD patients have been previously reported and such increase might be attributed to their biological roles. PCs and PEs are the most abundant glycerophospholipids in all eukaryotic cell membranes [13], whereas the biosynthesis of PCs in the human liver is mainly through the methylation of PEs [14]. Upon formation, hepatic PCs are specifically used to form monolayers of lipid droplets and can simultaneously function as a surfactant to prevent the monolayer from undergoing coalescence. Thus, larger lipid droplets are less likely to go through lipolysis in presence of abundant PCs, which eventually leads to liver steatosis [15]. Besides NAFLD, abnormal levels of PC and PE have also been observed in serum samples of patients with Alzheimer's disease and Parkinson's disease [16].
Fig. 4.
Heat map of hierarchical clustering of the serum samples from the control and NAFLD subjects. Red blocks indicate up-regulation, whereas blue indicates down-regulation (n = 8/group). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Abundance changes of representative serum metabolic and lipidomic biomarkers that differentiate the NAFLD and healthy subjects. (A) Comparison of lipids in the control and NAFLD subjects. (B) Comparison of acylcarnitines in the control and NAFLD subjects. (C) Comparison of organic acids in the control and NAFLD subjects. Bar plot represents the normalized intensity (normalized to TIC) of metabolites detected from each serum sample. Data are represented as normalized intensity ± SEM (n = 8/group). A comparison was made between the NAFLD and control subjects by Student's t-test. *, significantly different between the control and NAFLD subjects at p < 0.05; **, significantly different between the control and NAFLD subjects at p < 0.01.
LPC is a monoglycerophospholipid in which a phosphorylcholine moiety occupies a glycerol substitution site. An elevated level of LPCs was also noted in the sera of NAFLD patients compared with the healthy subjects. Previously, LPCs and DGs have been considered lipotoxic species and associated with hepatic injury [17]. Notably, LPC levels were also found elevated in a methionine-choline-deficient (MCD) diet-induced advanced NASH mouse model [18]. Thus, our results substantiate a possible link between increased LPC levels and NAFLD and further progression to NASH [19]. Additionally, the pathogenic role of DGs has been associated with its function as the activator of protein kinase C∊ (PKC∊), which leads to impaired insulin signaling. Thus, the high level of DGs in NAFLD patients, as we observed here, potentially explains why many NAFLD patients exhibit hepatic insulin resistance and type 2 diabetes [20]. Further, a previous study has recorded insulin-mediated deactivated expression of lipid hydrolysis enzymes in NAFLD patients [21], explaining the accumulation of DGs and MGs in sera of NAFLD patients, as detected in this study. Accordingly, we hypothesize that preventing the generation and accumulation of these lipotoxic lipids appears to serve as a therapeutic strategy for NAFLD treatment [24].
Besides lipids, our preliminary results also draw attention to acylcarnitines, such as octanoylcarnitine (C8), 3-hydroxy-cis-5-tetradecenoylcarnitine (C14), valerylcarnitine (C5), and two medium-chain acylcarnitines, hydroxyundecanoyl carnitine and hydroxylauroylcarnitine, that all exhibited marked increases in the NAFLD group compared to the healthy control group (Fig. 5B). We, thus, wondered why acylcarnitines were abnormally increased in NAFLD patients. Previous biochemical studies have shown that acylcarnitines derive from the esterification of L-carnitine in mammalian cells, a process responsible for transporting fatty acids into the mitochondria for subsequent β-oxidation [22]. Moreover, acylcarnitines, together with cardiolipin and ubiquinone, are lipid classes associated with mitochondrial function [23]. Thus, abnormal acylcarnitines levels in NAFLD patients could reflect the inability of mitochondria to properly metabolize fatty acids [24]. In agreement with this hypothesis, mitochondrial dysfunction has been suggested to play a pivotal role in the pathogenesis of NAFLD. The dysregulated homeostasis of acylcarnitines, as we detected, is supported by a recent study that records elevated octenoylcarnitine (C8), 3-hydroxy-cis-5-tetradecenoylcarnitine (C14), decanoylcarnitine (C10), dodecenoylcarnitine (C12) in serum samples of overweight NAFLD individuals [25]. Further, excessive total carnitine, including free carnitine, as well as short- and long-chain acylcarnitines, were also found in cirrhotic patients [26]. Together, our results confirm acylcarnitines as potential metabolic biomarkers of NAFLD, but how mitochondria dysfunction, as suggested by aberrant acylcarnitines levels, is correlated with NAFLD occurrence and progression warrants future investigations [27,29].
Besides lipids and acylcarnitines, we find organic acids hydroxydodecanedioic acid and deoxycholic acid 3-glucuronide are also useful metabolites in differentiating NAFLD and health subjects (Fig. 5C). Of note, deoxycholic acid 3-glucuronide in human serum is a natural metabolite of deoxycholic acid generated in the liver by uridine 5′-diphospho- glucuonyltransferase. Deoxycholic acid is a secondary bile acid produced by intestinal microbiota and its function is to emulsify fats for intestinal absorption. Interestingly, a recent study reported elevated serum concentrations of secondary bile acids in NAFLD patients [28], which may correlate with the increased deoxycholic acid 3-glucuronide, as we detected here. Together, the putative metabolic and lipidomic biomarkers we report in this pilot study suggest the potential to discover promising non-invasive histopathological markers for improving NAFLD diagnosis and stimulating therapeutic strategies.
Conclusions
Untargeted metabolomic analysis aims to comprehensively measure changes in endogenous metabolites. We have optimized a non-targeted serum metabolomic acquisition workflow using both T3 and HILIC columns and adopted both positive and negative ionization modes. Comparison of the four analytical modes has highlighted the necessity to include all modes of analysis in order to optimize coverage of the metabolome. Integration of the metabolomic data collected under the four combinatorial modes contributed to detecting 20 putative NAFLD biomarkers in this pilot study. The significant changes in lipid species, acylcarnitines, and organic acid levels from NAFLD patients revealed in this study agree with previous reports and reinforces the foundation for continued investigation of NAFLD biomarkers for translatation into diagnostics and therapeutic interventions in clinical practice.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the Natural Science Foundation of Jiangsu Province (BK20180079), Six Talent Peaks Project in Jiangsu Province (SWYY-101), International Industrial Technology Research Collaboration of Nanjing (201911008).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2021.10.001.
Contributor Information
Jun Wang, Email: wangjun868@126.com.
Hui Ye, Email: cpuyehui@cpu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Byrne C.D., Targher G. NAFLD: a multisystem disease. J. Hepatol. 2015;62(1):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., Wai-Sun Wong V., Yilmaz Y., George J., Fan J., Vos M.B. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology. 2019;69(6):2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 4.Wong V.-S., Adams L.A., de Lédinghen V., Wong G.-H., Sookoian S. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 2018;15(8):461–478. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 5.Gorden D.L., Myers D.S., Ivanova P.T., Fahy E., Maurya M.R., Gupta S., Min J., Spann N.J., McDonald J.G., Kelly S.L., Duan J., Sullards M.C., Leiker T.J., Barkley R.M., Quehenberger O., Armando A.M., Milne S.B., Mathews T.P., Armstrong M.D., Li C., Melvin W.V., Clements R.H., Washington M.K., Mendonsa A.M., Witztum J.L., Guan Z., Glass C.K., Murphy R.C., Dennis E.A., Merrill A.H., Russell D.W., Subramaniam S., Brown H.A. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 2015;56(3):722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokushige K., Hashimoto E., Kodama K., Tobari M., Matsushita N., Kogiso T., Taniai M., Torii N., Shiratori K., Nishizaki Y., Ohga T., Ohashi Y., Sato T. Serum metabolomic profile and potential biomarkers for severity of fibrosis in nonalcoholic fatty liver disease. J. Gastroenterol. 2013;48(12):1392–1400. doi: 10.1007/s00535-013-0766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morikawa H., Mano K., Horinaka H., Matsunaka T., Matsumoto Y., Ida T., Kawaguchi Y., Wada K., Kawada N. Development of non-invasive method for assessment of hepatic steatosis. Ultrasonics. 2016;72:195–200. doi: 10.1016/j.ultras.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Ioannou G.N., Nagana Gowda G.A., Djukovic D., Raftery D. Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites. 2020;10(4):168. doi: 10.3390/metabo10040168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S.-S., Long R., Song T., Zhang L.i., Dai Y.-L., Liu S.-W., Zhang P. UPLC-Q-TOF/MS Based Metabolomics Approach to Study the Hepatotoxicity of Cantharidin on Mice. Chem. Res. Toxicol. 2019;32(11):2204–2213. doi: 10.1021/acs.chemrestox.9b00233. [DOI] [PubMed] [Google Scholar]
- 10.Kalhan S.C., Guo L., Edmison J., Dasarathy S., McCullough A.J., Hanson R.W., Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60(3):404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai Y.-S., Chen W.-C., Kuo T.-C., Ho C.-T., Kuo C.-H., Tseng Y.J., Lu K.-H., Lin S.-H., Panyod S., Sheen L.-Y. Mass-Spectrometry-Based Serum Metabolomics of a C57BL/6J Mouse Model of High-Fat-Diet-Induced Non-alcoholic Fatty Liver Disease Development. J. Agric. Food Chem. 2015;63(35):7873–7884. doi: 10.1021/acs.jafc.5b02830. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari-Heckler S., Gan-Schreier H., Stremmel W., Chamulitrat W., Pathil A. Circulating Phospholipid Patterns in NAFLD Patients Associated with a Combination of Metabolic Risk Factors. Nutrients. 2018;10(5):649. doi: 10.3390/nu10050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perakakis N., Stefanakis K., Mantzoros C.S. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism. 2020;111:154320. doi: 10.1016/j.metabol.2020.154320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibellini F., Smith T.K. The Kennedy pathway--De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62(6):414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 15.Krahmer N., Guo Y.i., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H., Schmidt-Supprian M., Vance D., Mann M., Farese R., Walther T. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14(4):504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calzada E., Onguka O., Claypool S.M. Phosphatidylethanolamine Metabolism in Health and Disease. Int. Rev. Cell Mol. Biol. 2016;321:29–88. doi: 10.1016/bs.ircmb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kershaw E.E., Hamm J.K., Verhagen L.A.W., Peroni O., Katic M., Flier J.S. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55(1):148–157. [PMC free article] [PubMed] [Google Scholar]
- 18.Pathil A., Mueller J., Warth A., Chamulitrat W., Stremmel W. Ursodeoxycholyl lysophosphatidylethanolamide improves steatosis and inflammation in murine models of nonalcoholic fatty liver disease. Hepatology. 2012;55(5):1369–1378. doi: 10.1002/hep.25531. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan S.R., Butler L.M., Hoy A.J. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metab. 2021;9(1):2. doi: 10.1186/s40170-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen M.C., Shulman G.I. Roles of Diacylglycerols and Ceramides in Hepatic Insulin Resistance. Trends Pharmacol. Sci. 2017;38(7):649–665. doi: 10.1016/j.tips.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zechner R., Zimmermann R., Eichmann T., Kohlwein S., Haemmerle G., Lass A., Madeo F. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15(3):279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnoni A., Longo S., Gnoni G.V., Giudetti A.M. Carnitine in Human Muscle Bioenergetics: Can Carnitine Supplementation Improve Physical Exercise? Molecules. 2020;25(1):182. doi: 10.3390/molecules25010182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng K.-Y., Watt M.J., Rensen S., Greve J.W., Huynh K., Jayawardana K.S., Meikle P.J., Meex R.C.R. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res. 2018;59(10):1977–1986. doi: 10.1194/jlr.M085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjørndal B., et al. Associations between fatty acid oxidation, hepatic mitochondrial function, and plasma acylcarnitine levels in mice. Nutr Metab (Lond) 2018;15:10. doi: 10.1186/s12986-018-0241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M., Kim M., Kang M., Yoo H.J., Kim M.S., Ahn Y.-T., Sim J.-H., Jee S.H., Lee J.H. Effects of weight loss using supplementation with Lactobacillus strains on body fat and medium-chain acylcarnitines in overweight individuals. Food Funct. 2017;8(1):250–261. doi: 10.1039/c6fo00993j. [DOI] [PubMed] [Google Scholar]
- 26.Johnston D.G., Alberti K.G. Carbohydrate metabolism in liver disease. Clin. Endocrinol. Metab. 1976;5(3):675–702. doi: 10.1016/s0300-595x(76)80046-1. [DOI] [PubMed] [Google Scholar]
- 27.Li J., et al. Serum metabolomic analysis of the effect of exercise on nonalcoholic fatty liver disease. Endocr. Connect. 2019;8(4):299–308. doi: 10.1530/EC-19-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao N., et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67(10):1881–1891. doi: 10.1136/gutjnl-2017-314307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.