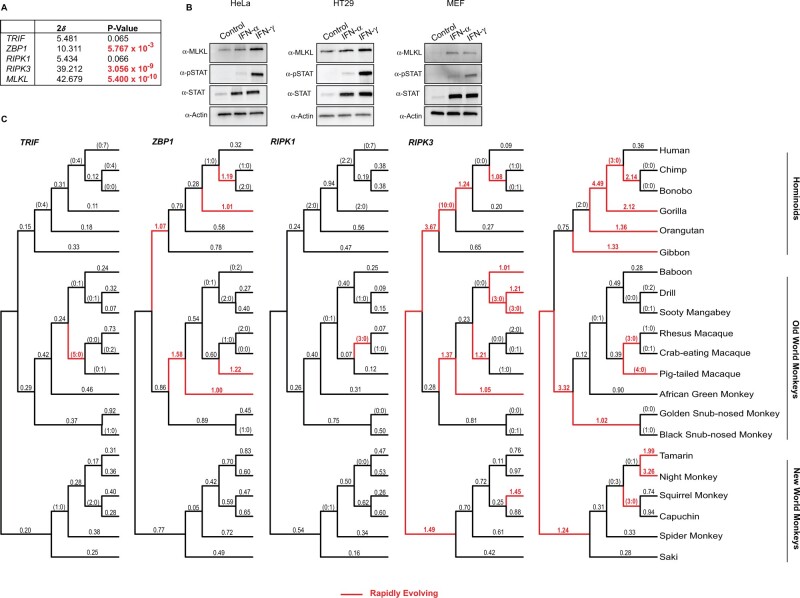
Fig. 2.
The rapid evolution of the necroptotic axis, RIPK3 and MLKL, across primate lineages. (A) Summary of rapid evolution results produced by NSsites analysis implemented in PAML for M7 × M8 (F3x4) Likelihood ratio test statistics (2δ) for necroptotic factors across a set of twenty-one matching primate species. (B) Western blot analysis demonstrates that MLKL protein is upregulated by IFNγ treatment in HeLa and HT29 cell lines and by IFNα as well as IFNγ treatment in murine embryonic fibroblasts. (C) dN/dS values estimated using FreeRatio analysis implemented in PAML across primate evolution. Rapidly evolving lineages (red branches) are defined by dN/dS >1, or greater than or equal to 3 nonsynonymous amino acid changes relative to zero synonymous amino acid changes.