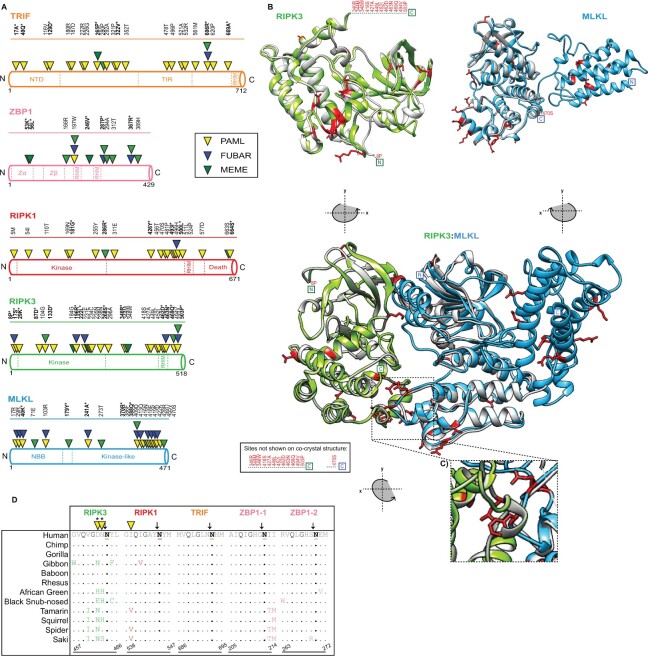
Fig. 3.
Widespread signatures of positive selection for cellular factors associated with necroptosis. (A) Rapidly evolving sites (triangles) identified—for primate TRIF (orange), ZBP1 (pink), RIPK1 (red), RIPK3 (green), and MLKL (blue)—were predicted using NSsites in PAML (yellow triangle), FUBAR (blue triangle) and MEME (green triangle). Sites with P-values <0.05 or posterior probabilities (P) >0.95 are listed above the protein cartoons, while sites with P-values <0.01 or posterior probabilities (P) >0.99 are also bolded and have an asterisk. Amino acid position refers to the human reference sequences. (B) Protein modeling for hRIPK3 and hMLKL highlights that multiple, discrete surfaces have rapid evolution signatures (red). H(uman)RIPK3 (green) homology model and the hMLKL (light blue) predicted structure (Murphy et al. 2013) are shown. The homology model was predicted using Swiss-Model (Waterhouse et al. 2018) and aligned to predicted structures m(ouse)RIPK3 (silver), mMLKL (silver), and mRIPK3: mMLKL (silver) (Xie et al. 2013). (C) Prominent rapid evolution (red) at the mRIPK3 hydrophobic pocket. (D) Rapid evolution exclusive to the RIPK3 RHIM domain proximal to the demonstrated EspL cleavage motif (Q*G**N—highlighted in black). Sequences spanning the EspL cleavage motif for 12 primates for RIPK3 (green), RIPK1 (red), TRIF (orange), and ZBP1 (pink) RHIM domains. The EspL cleavage site is underlined and displays a black arrow above the amino acid. Sites (triangles) with P-values <0.05 or posterior probabilities (P) >0.95 are listed above the protein cartoons. Sites with P-values <0.01 or posterior probabilities (P) >0.99 are bolded and have an asterisk. Amino acid positions refer to the human reference sequences. Note: PAML, MEME, and FUBAR analysis were performed for RIPK1 without drill monkey since this region was excluded due to trimming in preparation of sequences.