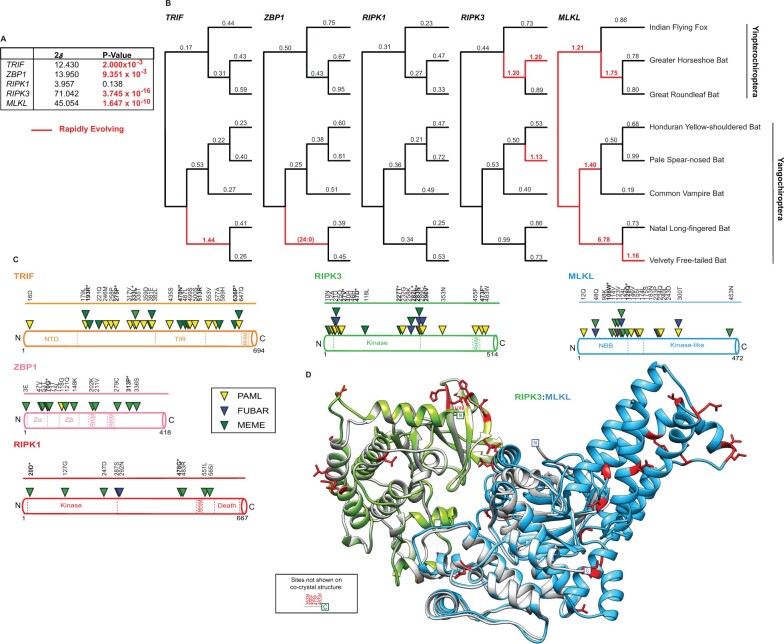
Fig. 4.
The rapid evolution of bat factors associated with necroptosis. (A) Summary of rapid evolution results produced using NSsites implemented in PAML for M7 × M8 (F3x4) Likelihood ratio test statistics (2δ) for necroptotic factors across a set of eight matching bat species. (B) dN/dS values estimated using Free Ratio analysis in PAML across bat phylogeny. Rapidly evolving lineages (red branches) defined by dN/dS >1, or greater than or equal to 3 nonsynonymous amino acid changes relative to zero synonymous amino acid changes. (C) Rapidly evolving sites (triangles) identified—for bat TRIF (orange), ZBP1 (pink), RIPK1 (red), RIPK3 (green), and MLKL (blue)—were predicted using NSsites in PAML (yellow triangle), FUBAR (blue triangle) and MEME (green triangle). Sites with P-values <0.05 or posterior probabilities (P) >0.95 are listed above the protein cartoons, while sites with P-values <0.01 or posterior probabilities (P) >0.99 are also bolded and have an asterisk. Amino acid alignment position refers to the Indian flying fox reference sequences. (D) Location of rapidly evolving sites (shown in red) on the b(at)RIPK3 (green) and bMLKL (light blue) homology models. The homology models were predicted using Swiss-Model (Waterhouse et al. 2018) and aligned to mRIPK3: mMLKL (silver) cocrystal structure (Xie et al. 2013).