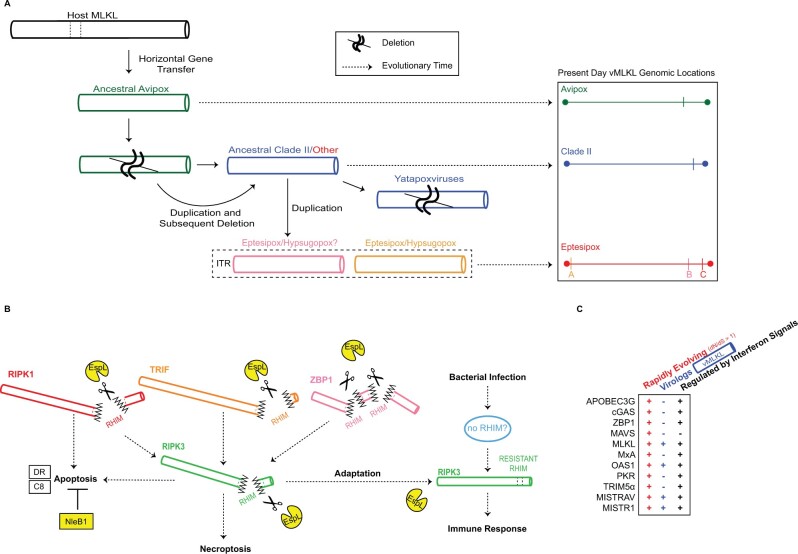
Fig. 9.
Model for pathogen-conflict driven evolution of viral and host MLKL. (A) Model for the acquisition and evolution of poxvirus MLKL. Data indicate that vMLKL was likely acquired by a vertebrate host presumably through a single horizontal gene transfer event to a shared poxvirus ancestor (fig. 7). This copy at the original site of integration, located on the right arm, has been maintained in Avipoxvirus (green) genomes to the present day (fig. 5). Following divergence from Avipoxvirus (figs. 6 and 7, supplementary fig. S5, Supplementary Material online), a duplication occurred followed by a deletion event in the common ancestor of Clade II (dark blue) and species classified as “Other” (red). This second copy also on the right arm is present in all Clade II species except in Yatapoxvirus lineages where it is lost (fig. 6). Subsequently, the second copy was duplicated to the ITR in the common ancestor of eptesipoxvirus and hypsugopoxvirus where it remains to present. While hypsugopoxvirus ITR sequence was not included in the final genome assembly, our synteny analysis (fig. 6) suggests vMLKL is also located on an ITR (dashed box) in this lineage. “ ” represents predicted deletion events. Dashed arrows represent the progression of evolutionary time (left to right). Scaled genome locations for present-day vMLKL copies are represented on the right. (B) Model for the adaptative escape of RIPK3 from EspL-like bacterial proteases supported by rapid evolutionary analysis. EspL cleaves RIPK1, RIPK3, TRIF and ZBP1 (Pearson et al. 2017). Through adaptation at the known EspL cleavage motif in the RHIM domain, RIPK3 may escape cleavage by EspL-like proteases to activate host immune responses. (C) MLKL displays all three key hallmarks that are often common to key host defense factors but as a subset, which OAS1 (Darby et al. 2014; Hancks et al. 2015; Mozzi et al. 2015), MISTRAV, and MISTR1 (Sorouri et al. 2020) also harbor. The factors noted are all rapidly evolving, possess a viral homolog/virolog, and are regulated by immune signals (includes upregulation and downregulation).
” represents predicted deletion events. Dashed arrows represent the progression of evolutionary time (left to right). Scaled genome locations for present-day vMLKL copies are represented on the right. (B) Model for the adaptative escape of RIPK3 from EspL-like bacterial proteases supported by rapid evolutionary analysis. EspL cleaves RIPK1, RIPK3, TRIF and ZBP1 (Pearson et al. 2017). Through adaptation at the known EspL cleavage motif in the RHIM domain, RIPK3 may escape cleavage by EspL-like proteases to activate host immune responses. (C) MLKL displays all three key hallmarks that are often common to key host defense factors but as a subset, which OAS1 (Darby et al. 2014; Hancks et al. 2015; Mozzi et al. 2015), MISTRAV, and MISTR1 (Sorouri et al. 2020) also harbor. The factors noted are all rapidly evolving, possess a viral homolog/virolog, and are regulated by immune signals (includes upregulation and downregulation).