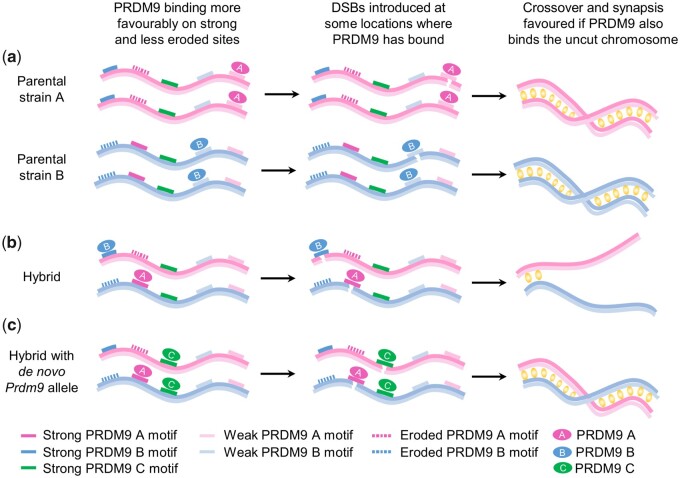
Fig. 1.
A model of the impact of PRDM9 binding on chromosome synapsis. (a) In two parental strains, A and B, different Prdm9 alleles are present, encoding PRDM9A and PRDM9B, which have distinct binding sites, with a spectrum of affinities (ranging from strong to weak). Due to the effects of mutation and recombination, these sites, preferentially the stronger sites, are frequently eroded. PRDM9 will still bind to its intact weaker binding sites and position DSBs to initiate the recombination process. PRDM9 is likely to bind at a similar level at these weaker sites at matched positions on the uncut homolog, enabling synapsis to occur and meiosis to proceed. (b) In the hybrid strain, two parental chromosomes and both PRDM9A and PRDM9B are present. The two PRDM9 variants now bind preferentially to the noneroded stronger binding sites on the complementary chromosome, leading to a reduction in cut sites at which PRDM9 is bound to the matched position on the homolog. Chromosome synapsis is inefficient and meiosis arrests. (c) In the hybrid, the replacement of one of the Prdm9 alleles with a de novo allele, Prdm9C encoding PRDM9C whose distinct binding sites are not eroded on either parental chromosome, leads to strong binding to this noneroded motif on both parental chromosomes, increasing the frequency of PRDM9 binding matched positions, favoring chromosome synapsis, and rescuing the meiotic arrest.